[English] 日本語
 Yorodumi
Yorodumi- PDB-3pse: Structure of a viral OTU domain protease bound to interferon-stim... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3pse | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a viral OTU domain protease bound to interferon-stimulated gene 15 (ISG15) | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | HYDROLASE/PROTEIN BINDING / viral deubiquitinase / 3-aminopropane / intein-mediated ligation / Crimean-Congo Hemorrhagic Fever virus / HYDROLASE-PROTEIN BINDING complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of protein oligomerization / ISG15-protein conjugation / RNA-templated viral transcription / regulation of type II interferon production / NS1 Mediated Effects on Host Pathways / protein localization to mitochondrion / response to type I interferon / negative stranded viral RNA replication / Modulation of host responses by IFN-stimulated genes / negative regulation of type I interferon-mediated signaling pathway ...positive regulation of protein oligomerization / ISG15-protein conjugation / RNA-templated viral transcription / regulation of type II interferon production / NS1 Mediated Effects on Host Pathways / protein localization to mitochondrion / response to type I interferon / negative stranded viral RNA replication / Modulation of host responses by IFN-stimulated genes / negative regulation of type I interferon-mediated signaling pathway / negative regulation of viral genome replication / RSV-host interactions / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / positive regulation of interleukin-10 production / protein deubiquitination / positive regulation of bone mineralization / endoplasmic reticulum unfolded protein response / ERAD pathway / negative regulation of protein ubiquitination / positive regulation of interferon-beta production / positive regulation of erythrocyte differentiation / integrin-mediated signaling pathway / Negative regulators of DDX58/IFIH1 signaling / Termination of translesion DNA synthesis / PKR-mediated signaling / DDX58/IFIH1-mediated induction of interferon-alpha/beta / modification-dependent protein catabolic process / integrin binding / ISG15 antiviral mechanism / response to virus / positive regulation of type II interferon production / protein tag activity / Interferon alpha/beta signaling / defense response to virus / symbiont-mediated suppression of host ISG15-protein conjugation / Hydrolases; Acting on ester bonds / symbiont-mediated perturbation of host ubiquitin-like protein modification / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / defense response to bacterium / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / innate immune response / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / ubiquitin protein ligase binding / DNA-templated transcription / SARS-CoV-2 activates/modulates innate and adaptive immune responses / extracellular region / nucleoplasm / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Crimean-Congo hemorrhagic fever virus Crimean-Congo hemorrhagic fever virus Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.3 Å MOLECULAR REPLACEMENT / Resolution: 2.3 Å | |||||||||
 Authors Authors | Bacik, J.P. / James, T.W. / Frias-Staheli, N. / Garcia-Sastre, A. / Mark, B.L. | |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2011 Journal: Proc.Natl.Acad.Sci.USA / Year: 2011Title: Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Authors: James, T.W. / Frias-Staheli, N. / Bacik, J.P. / Levingston Macleod, J.M. / Khajehpour, M. / Garcia-Sastre, A. / Mark, B.L. | |||||||||
| History |
| |||||||||
| Remark 650 | HELIX DETERMINATION METHOD: AUTHOR DETERMINED | |||||||||
| Remark 700 | SHEET DETERMINATION METHOD: AUTHOR DETERMINED |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3pse.cif.gz 3pse.cif.gz | 141.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3pse.ent.gz pdb3pse.ent.gz | 109.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3pse.json.gz 3pse.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ps/3pse https://data.pdbj.org/pub/pdb/validation_reports/ps/3pse ftp://data.pdbj.org/pub/pdb/validation_reports/ps/3pse ftp://data.pdbj.org/pub/pdb/validation_reports/ps/3pse | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3pt2SC  1z2mS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 19480.807 Da / Num. of mol.: 1 / Fragment: OTU domain (UNP residues 1-169) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Crimean-Congo hemorrhagic fever virus / Production host: Crimean-Congo hemorrhagic fever virus / Production host:  |
|---|---|
| #2: Protein | Mass: 17117.611 Da / Num. of mol.: 1 / Mutation: C78S Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ISG15, G1P2, UCRP / Production host: Homo sapiens (human) / Gene: ISG15, G1P2, UCRP / Production host:  |
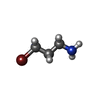
| #3: Chemical | ChemComp-4LJ / |
| #4: Chemical | ChemComp-GOL / |
| #5: Water | ChemComp-HOH / |
| Sequence details | S83N IS A NATURAL VARIANT |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.3 Å3/Da / Density % sol: 46.61 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6.4 Details: 100 mM MES, 23% PEG6000, pH 6.4, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CLSI CLSI  / Beamline: 08ID-1 / Wavelength: 0.9795 Å / Beamline: 08ID-1 / Wavelength: 0.9795 Å |
| Detector | Type: RAYONIX MX-300 / Detector: CCD / Date: Feb 6, 2010 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→34.9 Å / Num. all: 14673 / Num. obs: 14495 / % possible obs: 97.8 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 2.5 / Redundancy: 3.1 % / Rmerge(I) obs: 0.107 / Rsym value: 0.129 / Net I/σ(I): 7.4 |
| Reflection shell | Resolution: 2.3→2.42 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.566 / Mean I/σ(I) obs: 2.3 / Rsym value: 0.68 / % possible all: 98 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRIES 1Z2M AND 3PT2 Resolution: 2.3→34.671 Å / SU ML: 0.28 / σ(F): 1 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.83 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 46.22 Å2 / ksol: 0.359 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→34.671 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj