[English] 日本語
 Yorodumi
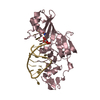
Yorodumi- PDB-3isw: Crystal structure of filamin-A immunoglobulin-like repeat 21 boun... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3isw | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of filamin-A immunoglobulin-like repeat 21 bound to an N-terminal peptide of CFTR | ||||||
 Components Components |
| ||||||
 Keywords Keywords | STRUCTURAL PROTEIN / protein-peptide complex / Acetylation / Actin-binding / Alternative splicing / Cytoplasm / Cytoskeleton / Disease mutation / Phosphoprotein / Polymorphism / ATP-binding / Chloride / Chloride channel / Glycoprotein / Hydrolase / Ion transport / Ionic channel / Membrane / Nucleotide-binding / Transmembrane / Transport | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of membrane repolarization during atrial cardiac muscle cell action potential / regulation of membrane repolarization during cardiac muscle cell action potential / establishment of Sertoli cell barrier / Myb complex / glycoprotein Ib-IX-V complex / adenylate cyclase-inhibiting dopamine receptor signaling pathway / formation of radial glial scaffolds / positive regulation of voltage-gated chloride channel activity / positive regulation of integrin-mediated signaling pathway / : ...regulation of membrane repolarization during atrial cardiac muscle cell action potential / regulation of membrane repolarization during cardiac muscle cell action potential / establishment of Sertoli cell barrier / Myb complex / glycoprotein Ib-IX-V complex / adenylate cyclase-inhibiting dopamine receptor signaling pathway / formation of radial glial scaffolds / positive regulation of voltage-gated chloride channel activity / positive regulation of integrin-mediated signaling pathway / : / Sec61 translocon complex binding / channel-conductance-controlling ATPase / intracellularly ATP-gated chloride channel activity / positive regulation of enamel mineralization / transepithelial water transport / RHO GTPases regulate CFTR trafficking / actin crosslink formation / blood coagulation, intrinsic pathway / tubulin deacetylation / amelogenesis / intracellular pH elevation / OAS antiviral response / positive regulation of actin filament bundle assembly / positive regulation of neuron migration / chloride channel inhibitor activity / : / protein localization to bicellular tight junction / multicellular organismal-level water homeostasis / Golgi-associated vesicle membrane / Fc-gamma receptor I complex binding / Cell-extracellular matrix interactions / cholesterol transport / apical dendrite / positive regulation of potassium ion transmembrane transport / positive regulation of platelet activation / positive regulation of neural precursor cell proliferation / bicarbonate transport / bicarbonate transmembrane transporter activity / membrane hyperpolarization / chloride channel regulator activity / protein localization to cell surface / vesicle docking involved in exocytosis / wound healing, spreading of cells / chloride transmembrane transporter activity / podosome / negative regulation of transcription by RNA polymerase I / megakaryocyte development / GP1b-IX-V activation signalling / cholesterol biosynthetic process / SMAD binding / receptor clustering / sperm capacitation / chloride channel activity / cortical cytoskeleton / RHOQ GTPase cycle / RHO GTPases activate PAKs / semaphorin-plexin signaling pathway / positive regulation of exocytosis / cilium assembly / ATPase-coupled transmembrane transporter activity / mitotic spindle assembly / potassium channel regulator activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / ABC-type transporter activity / chloride channel complex / : / 14-3-3 protein binding / release of sequestered calcium ion into cytosol / positive regulation of substrate adhesion-dependent cell spreading / protein sequestering activity / cellular response to forskolin / regulation of cell migration / chloride transmembrane transport / cellular response to cAMP / response to endoplasmic reticulum stress / dendritic shaft / protein localization to plasma membrane / PDZ domain binding / actin filament / establishment of protein localization / clathrin-coated endocytic vesicle membrane / establishment of localization in cell / mRNA transcription by RNA polymerase II / Defective CFTR causes cystic fibrosis / Late endosomal microautophagy / G protein-coupled receptor binding / recycling endosome / cerebral cortex development / negative regulation of protein catabolic process / positive regulation of protein import into nucleus / ABC-family proteins mediated transport / transmembrane transport / small GTPase binding / platelet aggregation / kinase binding / Z disc / recycling endosome membrane / Chaperone Mediated Autophagy / Aggrephagy / cell-cell junction Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.8 Å MOLECULAR REPLACEMENT / Resolution: 2.8 Å | ||||||
 Authors Authors | Xu, Z. / Page, R. / Qin, J. / Ithychanda, S.S. / Liu, J.M. / Misra, S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2010 Journal: J.Biol.Chem. / Year: 2010Title: Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin. Authors: Smith, L. / Page, R.C. / Xu, Z. / Kohli, E. / Litman, P. / Nix, J.C. / Ithychanda, S.S. / Liu, J. / Qin, J. / Misra, S. / Liedtke, C.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3isw.cif.gz 3isw.cif.gz | 51.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3isw.ent.gz pdb3isw.ent.gz | 36.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3isw.json.gz 3isw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  3isw_validation.pdf.gz 3isw_validation.pdf.gz | 442.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  3isw_full_validation.pdf.gz 3isw_full_validation.pdf.gz | 444.7 KB | Display | |
| Data in XML |  3isw_validation.xml.gz 3isw_validation.xml.gz | 9.5 KB | Display | |
| Data in CIF |  3isw_validation.cif.gz 3isw_validation.cif.gz | 12.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/is/3isw https://data.pdbj.org/pub/pdb/validation_reports/is/3isw ftp://data.pdbj.org/pub/pdb/validation_reports/is/3isw ftp://data.pdbj.org/pub/pdb/validation_reports/is/3isw | HTTPS FTP |
-Related structure data
| Related structure data |  2brqS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | THE BIOLOGICAL ASSEMBLY IS THE DIMERIC COMPLEX OF THE PROTEIN WITH THE PEPTIDE, REPRESENTED BY THE ENTIRE CONTENTS OF THE ASYMMETRIC UNIT. THE SOFTWARE-GENERATED WORD 'TRIMERIC' IN REMARK 350 IS A POLYMERIC COUNT. |
- Components
Components
| #1: Protein | Mass: 10177.196 Da / Num. of mol.: 2 / Fragment: UNP residues 2236-2329 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FLN, FLN-21, FLN1, FLNA / Production host: Homo sapiens (human) / Gene: FLN, FLN-21, FLN1, FLNA / Production host:  #2: Protein/peptide | | Mass: 2009.345 Da / Num. of mol.: 1 / Fragment: UNP residues 5-22 / Mutation: R21A / Source method: obtained synthetically Details: Synthetic peptide based on the sequence 5-22 of Homo sapiens CFTR (UniProt entry P13569, CFTR_HUMAN) References: UniProt: P13569 #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.13 Å3/Da / Density % sol: 76.02 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 0.1M Bis-Tris Propane pH 7.0, 60% Tacsimate, VAPOR DIFFUSION, HANGING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 4.2.2 / Wavelength: 1 Å / Beamline: 4.2.2 / Wavelength: 1 Å |
| Detector | Type: NOIR-1 / Detector: CCD / Date: Jul 1, 2007 Details: ROSENBAUM-ROCK MONOCHROMATOR FOR SAGITAL FOCUSING, ROSENBAUM-ROCK VERTICAL FOCUSING MIRROR |
| Radiation | Monochromator: double crystal / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.8→42.97 Å / Num. all: 13108 / Num. obs: 12452 / % possible obs: 96.6 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 5 / Redundancy: 8.53 % / Biso Wilson estimate: 75.7 Å2 / Rmerge(I) obs: 0.071 / Rsym value: 0.071 / Χ2: 0.94 / Net I/σ(I): 13.8 / Scaling rejects: 3169 |
| Reflection shell | Resolution: 2.8→2.9 Å / Redundancy: 10.41 % / Rmerge(I) obs: 0.472 / Mean I/σ(I) obs: 3.3 / Num. unique all: 1177 / Rsym value: 0.472 / Χ2: 1.34 / % possible all: 98.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 2BRQ Resolution: 2.8→34.73 Å / Cor.coef. Fo:Fc: 0.919 / Cor.coef. Fo:Fc free: 0.907 / Occupancy max: 1 / Occupancy min: 1 / SU B: 14.066 / SU ML: 0.277 / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / ESU R: 0.403 / ESU R Free: 0.313 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 104.74 Å2 / Biso mean: 69.191 Å2 / Biso min: 37.96 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→34.73 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.8→2.873 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj











