[English] 日本語
 Yorodumi
Yorodumi- PDB-3a3h: CELLOTRIOSE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGAR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3a3h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CELLOTRIOSE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHERANS AT 1.6 A RESOLUTION | |||||||||
 Components Components | ENDOGLUCANASE | |||||||||
 Keywords Keywords | HYDROLASE / CELLULOSE DEGRADATION / ENDOGLUCANASE / GLYCOSIDE HYDROLASE FAMILY 5 | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellulase / cellulase activity / cellulose catabolic process / carbohydrate binding / extracellular region Similarity search - Function | |||||||||
| Biological species |  Bacillus agaradhaerens (bacteria) Bacillus agaradhaerens (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / ISOMORPHOUS WITH NATIVE STRUCTURE / Resolution: 1.64 Å X-RAY DIFFRACTION / ISOMORPHOUS WITH NATIVE STRUCTURE / Resolution: 1.64 Å | |||||||||
 Authors Authors | Davies, G.J. / Brzozowski, A.M. / Andersen, K. / Schulein, M. | |||||||||
 Citation Citation |  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase. Authors: Davies, G.J. / Mackenzie, L. / Varrot, A. / Dauter, M. / Brzozowski, A.M. / Schulein, M. / Withers, S.G. #1:  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Structure of the Bacillus Agaradherans Family 5 Endoglucanase at 1.6 A and its Cellobiose Complex at 2.0 A Resolution Authors: Davies, G.J. / Dauter, M. / Brzozowski, A.M. / Bjornvad, M.E. / Andersen, K.V. / Schulein, M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3a3h.cif.gz 3a3h.cif.gz | 82.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3a3h.ent.gz pdb3a3h.ent.gz | 60.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3a3h.json.gz 3a3h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/a3/3a3h https://data.pdbj.org/pub/pdb/validation_reports/a3/3a3h ftp://data.pdbj.org/pub/pdb/validation_reports/a3/3a3h ftp://data.pdbj.org/pub/pdb/validation_reports/a3/3a3h | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 33653.746 Da / Num. of mol.: 1 / Fragment: CATALYTIC CORE Source method: isolated from a genetically manipulated source Details: THIS IS A COMPLEX WITH B-D-CELLOTRIOSE BOUND IN THE -1, -2 AND -3 SITES OF THE ENZYME Source: (gene. exp.)  Bacillus agaradhaerens (bacteria) / Strain: AC13 / Plasmid: THERMAMYL-AMYLASE PROMOTER SYSTEM / Production host: Bacillus agaradhaerens (bacteria) / Strain: AC13 / Plasmid: THERMAMYL-AMYLASE PROMOTER SYSTEM / Production host:  |
|---|---|
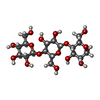
| #2: Polysaccharide | beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose / alpha-cellotriose |
| #3: Water | ChemComp-HOH / |
| Compound details | THE FIRST 3 RESIDUES ARE DISORDERED SO IT STARTS WITH RESIDUE SER 4. THIS THE NATURALLY OCCURRING ...THE FIRST 3 RESIDUES ARE DISORDERED |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 44 % | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 4.5 / Details: pH 4.5 | |||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / Details: Davies, G.J., (1998) Biochemistry, 37, 1926. | |||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: May 1, 1997 / Details: LONG FOCUSSING MIRRORS (MSC) |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.64→20 Å / Num. obs: 19752 / % possible obs: 99.5 % / Redundancy: 3.2 % / Biso Wilson estimate: 13 Å2 / Rmerge(I) obs: 0.045 / Rsym value: 0.045 / Net I/σ(I): 19.1 |
| Reflection shell | Resolution: 1.64→1.7 Å / Redundancy: 2.9 % / Rmerge(I) obs: 0.174 / Mean I/σ(I) obs: 7.6 / Rsym value: 0.174 / % possible all: 98.8 |
| Reflection | *PLUS Lowest resolution: 15 Å / Redundancy: 4.2 % / Rmerge(I) obs: 0.046 |
| Reflection shell | *PLUS % possible obs: 97.1 % / Redundancy: 4.2 % / Rmerge(I) obs: 0.184 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: ISOMORPHOUS WITH NATIVE STRUCTURE Resolution: 1.64→15 Å / Cross valid method: THROUGHOUT / σ(F): 0 Details: ESTIMATED COORDINATE ERROR. ESD FROM SIGMAA (A) : 0.013 LOW RESOLUTION CUTOFF (A) : 15
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 12.6 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati d res low obs: 15 Å / Luzzati sigma a obs: 0.01 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.64→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.148 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller
















 PDBj
PDBj