[English] 日本語
 Yorodumi
Yorodumi- PDB-2y3a: Crystal structure of p110beta in complex with icSH2 of p85beta an... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2y3a | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSFERASE / PHOSPHOINOSITIDE 3-KINASE / RTK | ||||||
| Function / homology |  Function and homology information Function and homology informationRHOJ GTPase cycle / IRS-mediated signalling / Signaling by ALK / Signaling by LTK / Tie2 Signaling / RND3 GTPase cycle / PI3K/AKT activation / PI3K Cascade / Role of LAT2/NTAL/LAB on calcium mobilization / RHOF GTPase cycle ...RHOJ GTPase cycle / IRS-mediated signalling / Signaling by ALK / Signaling by LTK / Tie2 Signaling / RND3 GTPase cycle / PI3K/AKT activation / PI3K Cascade / Role of LAT2/NTAL/LAB on calcium mobilization / RHOF GTPase cycle / RHOU GTPase cycle / RAC3 GTPase cycle / Downstream signal transduction / CDC42 GTPase cycle / RAC2 GTPase cycle / GPVI-mediated activation cascade / Interleukin-7 signaling / : / Signaling by SCF-KIT / negative regulation of sprouting angiogenesis / RND2 GTPase cycle / RND1 GTPase cycle / Role of phospholipids in phagocytosis / negative regulation of vascular endothelial growth factor signaling pathway / Co-stimulation by ICOS / RAC1 GTPase cycle / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / CD28 dependent PI3K/Akt signaling / RHOB GTPase cycle / Interleukin receptor SHC signaling / Synthesis of PIPs at the plasma membrane / Extra-nuclear estrogen signaling / G alpha (q) signalling events / positive regulation of neutrophil apoptotic process / PIP3 activates AKT signaling / RHOA GTPase cycle / Downstream TCR signaling / Regulation of signaling by CBL / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / RAF/MAP kinase cascade / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / regulation of cell-matrix adhesion / Interleukin-3, Interleukin-5 and GM-CSF signaling / RET signaling / regulation of actin filament polymerization / phosphatidylinositol 3-kinase complex / phosphatidylinositol 3-kinase regulatory subunit binding / embryonic cleavage / VEGFA-VEGFR2 Pathway / regulation of stress fiber assembly / 1-phosphatidylinositol-4-phosphate 3-kinase activity / sphingosine-1-phosphate receptor signaling pathway / phosphatidylinositol 3-kinase complex, class IA / endothelial cell proliferation / phosphatidylinositol-3-phosphate biosynthetic process / DAP12 signaling / angiogenesis involved in wound healing / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / phosphatidylinositol-4,5-bisphosphate 3-kinase / 1-phosphatidylinositol-3-kinase activity / natural killer cell mediated cytotoxicity / phosphatidylinositol-mediated signaling / phosphatidylinositol phosphate biosynthetic process / homophilic cell-cell adhesion / intracellular glucose homeostasis / positive regulation of Rac protein signal transduction / insulin receptor substrate binding / regulation of protein localization to plasma membrane / negative regulation of MAPK cascade / phosphotyrosine residue binding / positive regulation of endothelial cell migration / positive regulation of cell adhesion / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / response to endoplasmic reticulum stress / response to ischemia / phosphatidylinositol 3-kinase/protein kinase B signal transduction / brush border membrane / positive regulation of protein import into nucleus / platelet activation / receptor tyrosine kinase binding / regulation of autophagy / autophagy / cellular response to insulin stimulus / endocytosis / intracellular calcium ion homeostasis / positive regulation of nitric oxide biosynthetic process / insulin receptor signaling pathway / cell migration / protein transport / midbody / protein phosphatase binding / non-specific serine/threonine protein kinase / protein heterodimerization activity / protein serine kinase activity / focal adhesion / protein serine/threonine kinase activity / positive regulation of gene expression / nucleolus / positive regulation of transcription by RNA polymerase II / nucleoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.3 Å MOLECULAR REPLACEMENT / Resolution: 3.3 Å | ||||||
 Authors Authors | Zhang, X. / Vadas, O. / Perisic, O. / Williams, R.L. | ||||||
 Citation Citation |  Journal: Mol.Cell / Year: 2011 Journal: Mol.Cell / Year: 2011Title: Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism. Authors: Zhang, X. / Vadas, O. / Perisic, O. / Anderson, K.E. / Clark, J. / Hawkins, P.T. / Stephens, L.R. / Williams, R.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2y3a.cif.gz 2y3a.cif.gz | 261.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2y3a.ent.gz pdb2y3a.ent.gz | 205.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2y3a.json.gz 2y3a.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/y3/2y3a https://data.pdbj.org/pub/pdb/validation_reports/y3/2y3a ftp://data.pdbj.org/pub/pdb/validation_reports/y3/2y3a ftp://data.pdbj.org/pub/pdb/validation_reports/y3/2y3a | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2whgS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 125242.070 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Protein | Mass: 35346.574 Da / Num. of mol.: 1 / Fragment: ICSH2 DOMAIN, RESIDUES 423-722 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
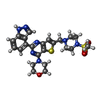
| #3: Chemical | ChemComp-GD9 / |
| Sequence details | N-TERMINAL 6XHIS TAG |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.48 Å3/Da / Density % sol: 64.71 % / Description: NONE |
|---|---|
| Crystal grow | pH: 6 Details: PROTEIN WAS CRYSTALLIZED FROM 12% PEG 3350, 0.1 M POTASSIUM CITRATE PH 6, 0.4 M LITHIUM SULFATE |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.9765 / Beamline: ID14-4 / Wavelength: 0.9765 |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Dec 5, 2009 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9765 Å / Relative weight: 1 |
| Reflection | Resolution: 3.3→44 Å / Num. obs: 34250 / % possible obs: 98.9 % / Observed criterion σ(I): 1 / Redundancy: 5.2 % / Biso Wilson estimate: 90.8 Å2 / Rmerge(I) obs: 0.1 / Net I/σ(I): 10.4 |
| Reflection shell | Resolution: 3.3→3.5 Å / Redundancy: 5.4 % / Rmerge(I) obs: 0.67 / Mean I/σ(I) obs: 1.9 / % possible all: 99.9 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2WHG Resolution: 3.3→44.68 Å / Cor.coef. Fo:Fc: 0.8743 / Cor.coef. Fo:Fc free: 0.8398 / Cross valid method: THROUGHOUT / σ(F): 0 Details: RESIDUES 1-12, 228-234, 299-3 -1064 ARE DISORDERED.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 127.72 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.906 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.3→44.68 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.3→3.4 Å / Total num. of bins used: 17
|
 Movie
Movie Controller
Controller










 PDBj
PDBj