[English] 日本語
 Yorodumi
Yorodumi- PDB-2wwd: 3D-structure of the modular autolysin LytC from Streptococcus pne... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2wwd | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | ||||||||||||
 Components Components | 1,4-BETA-N-ACETYLMURAMIDASE | ||||||||||||
 Keywords Keywords | HYDROLASE / GLYCOSIDASE / CHOLINE-BINDING PROTEINS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidoglycan beta-N-acetylmuramidase / peptidoglycan beta-N-acetylmuramidase activity / mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase / mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase activity / carbohydrate catabolic process / peptidoglycan catabolic process / cell wall macromolecule catabolic process / lysozyme / lysozyme activity Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.25 Å MOLECULAR REPLACEMENT / Resolution: 2.25 Å | ||||||||||||
 Authors Authors | Perez-Dorado, I. / Sanles, R. / Hermoso, J.A. / Gonzalez, A. / Garcia, A. / Garcia, P. / Garcia, J.L. | ||||||||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2010 Journal: Nat.Struct.Mol.Biol. / Year: 2010Title: Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc. Authors: Perez-Dorado, I. / Gonzalez, A. / Morales, M. / Sanles, R. / Striker, W. / Vollmer, W. / Mobashery, S. / Garcia, J.L. / Martinez-Ripoll, M. / Garcia, P. / Hermoso, J.A. #1: Journal: J.Biol.Chem. / Year: 2008 Title: Insights Into the Structure-Function Relationships of Pneumococcal Cell Wall Lysozymes, Lytc and Cpl- 1. Authors: Monterroso, B. / Saiz, J.L. / Garcia, P. / Garcia, J.L. / Menendez, M. #2: Journal: Biochem.J. / Year: 2005 Title: Unravelling the Structure of the Pneumococcal Autolytic Lysozyme. Authors: Monterroso, B. / Lopez-Zumel, C. / Garcia, J.L. / Saiz, J.L. / Garcia, P. / Campillo, N.E. / Menendez, M. #3: Journal: Mol.Microbiol. / Year: 1999 Title: The Molecular Characterization of the First Autolytic Lysozyme of Streptococcus Pneumoniae Reveals Evolutionary Mobile Domains. Authors: Garcia, P. / Paz Gonzalez, M. / Garcia, E. / Garcia, J.L. / Lopez, R. | ||||||||||||
| History |
| ||||||||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AL" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AL" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 8-STRANDED BARREL THIS IS REPRESENTED BY A 9-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2wwd.cif.gz 2wwd.cif.gz | 112.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2wwd.ent.gz pdb2wwd.ent.gz | 84.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2wwd.json.gz 2wwd.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ww/2wwd https://data.pdbj.org/pub/pdb/validation_reports/ww/2wwd ftp://data.pdbj.org/pub/pdb/validation_reports/ww/2wwd ftp://data.pdbj.org/pub/pdb/validation_reports/ww/2wwd | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2ww5SC  2wwcC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Sugars , 2 types, 2 molecules A
| #1: Protein | Mass: 55270.633 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid Source method: isolated from a genetically manipulated source |
-Non-polymers , 5 types, 152 molecules 
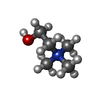







| #3: Chemical | ChemComp-GOL / #4: Chemical | ChemComp-CHT / #5: Chemical | ChemComp-ALA / | #6: Chemical | ChemComp-GLN / | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Sequence details | ENTRY ACCESSION CODE CORRESPONDS TO THE IMMATURE PROTEIN WHICH POSSESSES A SIGNAL PEPTIDE OF 33 ...ENTRY ACCESSION CODE CORRESPOND |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.74 Å3/Da / Density % sol: 55.1 % / Description: NONE |
|---|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-4 / Wavelength: 0.9795 / Beamline: ID14-4 / Wavelength: 0.9795 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Jul 14, 2008 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 2.25→75 Å / Num. obs: 27909 / % possible obs: 99.4 % / Observed criterion σ(I): 0 / Redundancy: 3.5 % / Rmerge(I) obs: 0.08 / Net I/σ(I): 12.7 |
| Reflection shell | Resolution: 2.25→2.37 Å / Redundancy: 3.6 % / Rmerge(I) obs: 0.5 / Mean I/σ(I) obs: 2.6 / % possible all: 99.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2WW5 Resolution: 2.25→57.17 Å / Cor.coef. Fo:Fc: 0.947 / Cor.coef. Fo:Fc free: 0.931 / SU B: 12.134 / SU ML: 0.153 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.291 / ESU R Free: 0.208 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: THE FIRST 37 AA OF THE POLYPEPTIDE CHAIN WERE NOT MODELLED DUE TO POOR ELECTRON DENSITY. ATOM RECORD CONTAINS RESIDUAL B FACTORS ONLY.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.15 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.25→57.17 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj



