[English] 日本語
 Yorodumi
Yorodumi- PDB-2v3v: A New Catalytic Mechanism of Periplasmic Nitrate Reductase from D... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2v3v | ||||||
|---|---|---|---|---|---|---|---|
| Title | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | ||||||
 Components Components | PERIPLASMIC NITRATE REDUCTASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / NITRATE ASSIMILATION / NITROGENOUS ACCEPTOR / ELECTRON TRANSPORT / DISSIMILATORY NITRATE REDUCTASE / MOLYBDOPTERIN COFACTOR / SULFIDO LIGAND | ||||||
| Function / homology |  Function and homology information Function and homology informationnitrate reductase (cytochrome) / nitrate reductase (cytochrome) activity / nitrate reductase complex / molybdenum ion binding / Mo-molybdopterin cofactor biosynthetic process / molybdopterin cofactor binding / nitrate assimilation / 4 iron, 4 sulfur cluster binding / electron transfer activity / periplasmic space ...nitrate reductase (cytochrome) / nitrate reductase (cytochrome) activity / nitrate reductase complex / molybdenum ion binding / Mo-molybdopterin cofactor biosynthetic process / molybdopterin cofactor binding / nitrate assimilation / 4 iron, 4 sulfur cluster binding / electron transfer activity / periplasmic space / iron ion binding / membrane Similarity search - Function | ||||||
| Biological species |  DESULFOVIBRIO DESULFURICANS (bacteria) DESULFOVIBRIO DESULFURICANS (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.99 Å MOLECULAR REPLACEMENT / Resolution: 1.99 Å | ||||||
 Authors Authors | Najmudin, S. / Gonzalez, P.J. / Trincao, J. / Coelho, C. / Mukhopadhyay, A. / Romao, C.C. / Moura, I. / Moura, J.J. / Brondino, C.D. / Romao, M.J. | ||||||
 Citation Citation |  Journal: J.Biol.Inorg.Chem. / Year: 2008 Journal: J.Biol.Inorg.Chem. / Year: 2008Title: Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum. Authors: Najmudin, S. / Gonzalez, P.J. / Trincao, J. / Coelho, C. / Mukhopadhyay, A. / Cerqueira, N.M.F.S.A. / Romao, C.C. / Moura, I. / Moura, J.J.G. / Brondino, C.D. / Romao, M.J. #1: Journal: J.Biol.Inorg.Chem. / Year: 2006 Title: Epr and Redox Properties of Periplasmic Nitrate Reductase from Desulfovibrio Desulfuricans Atcc 27774 Authors: Gonzalez, P.J. / Rivas, M.G. / Brondino, C.D. / Bursakov, S.A. / Moura, I. / Moura, J.J.G. | ||||||
| History |
| ||||||
| Remark 700 | SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AG" IN EACH CHAIN ON SHEET RECORDS BELOW ... SHEET DETERMINATION METHOD: DSSP THE SHEETS PRESENTED AS "AG" IN EACH CHAIN ON SHEET RECORDS BELOW IS ACTUALLY AN 6-STRANDED BARREL THIS IS REPRESENTED BY A 7-STRANDED SHEET IN WHICH THE FIRST AND LAST STRANDS ARE IDENTICAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2v3v.cif.gz 2v3v.cif.gz | 177.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2v3v.ent.gz pdb2v3v.ent.gz | 137.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2v3v.json.gz 2v3v.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v3/2v3v https://data.pdbj.org/pub/pdb/validation_reports/v3/2v3v ftp://data.pdbj.org/pub/pdb/validation_reports/v3/2v3v ftp://data.pdbj.org/pub/pdb/validation_reports/v3/2v3v | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2jimC  2jioC  2jipC  2jiqC  2jirC  2v45C  2napS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 80393.805 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  DESULFOVIBRIO DESULFURICANS (bacteria) / References: UniProt: P81186, nitrate reductase DESULFOVIBRIO DESULFURICANS (bacteria) / References: UniProt: P81186, nitrate reductase |
|---|
-Non-polymers , 6 types, 766 molecules 



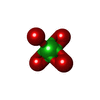






| #2: Chemical | ChemComp-SF4 / | ||||||
|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-MO / | ||||||
| #4: Chemical | | #5: Chemical | ChemComp-UNX / | #6: Chemical | ChemComp-LCP / #7: Water | ChemComp-HOH / | |
-Details
| Nonpolymer details | THE RESIDUE A 813 WAS ORIGINALLY DEPOSITED AS A ISOLATED SULFUR ATOM. AS PART OF REMEDIATION, THIS ...THE RESIDUE A 813 WAS ORIGINALLY |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.42 Å3/Da / Density % sol: 55.44 % / Description: NONE |
|---|---|
| Crystal grow | pH: 6.5 Details: 0.1M MES, PH 6.5, 7.5% PEG 8000 A COCRYSTALLISATION WITH 10MM OF KCLO4 (PERCHLORATE) IN 200MM OF DDNAPA (IE PROTEIN) WHICH HAD BEEN REDUCED BY DITHIONITE IN ANAEROBIC CONDITIONS AND THEN AIR ...Details: 0.1M MES, PH 6.5, 7.5% PEG 8000 A COCRYSTALLISATION WITH 10MM OF KCLO4 (PERCHLORATE) IN 200MM OF DDNAPA (IE PROTEIN) WHICH HAD BEEN REDUCED BY DITHIONITE IN ANAEROBIC CONDITIONS AND THEN AIR REOXIDISED BEFORE SETTTING UP FOR CRYSTALLISATIONS STRAIGHT AFTER THAWING FROM -80C. |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-3 / Wavelength: 0.931 / Beamline: ID14-3 / Wavelength: 0.931 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Nov 22, 2004 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.931 Å / Relative weight: 1 |
| Reflection | Resolution: 1.91→46.13 Å / Num. obs: 454081 / % possible obs: 93.5 % / Observed criterion σ(I): 1 / Redundancy: 7 % / Rmerge(I) obs: 0.114 / Net I/σ(I): 10 |
| Reflection shell | Resolution: 1.91→2.01 Å / Redundancy: 2.6 % / Rmerge(I) obs: 1.33 / Mean I/σ(I) obs: 0.8 / % possible all: 60.4 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2NAP Resolution: 1.99→92.45 Å / Cor.coef. Fo:Fc: 0.96 / Cor.coef. Fo:Fc free: 0.932 / SU B: 9.545 / SU ML: 0.144 / TLS residual ADP flag: UNVERIFIED / Cross valid method: THROUGHOUT / ESU R: 0.17 / ESU R Free: 0.17 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. THE FIRST 3 RESIDUES ARE DISORDERED.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 19.55 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.99→92.45 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








