[English] 日本語
 Yorodumi
Yorodumi- PDB-2usn: CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2usn | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-141803 | ||||||
 Components Components | STROMELYSIN-1 | ||||||
 Keywords Keywords | HYDROLASE / METALLOPROTEASE / FIBROBLAST / COLLAGEN DEGRADATION | ||||||
| Function / homology |  Function and homology information Function and homology informationstromelysin 1 / cellular response to UV-A / regulation of neuroinflammatory response / Assembly of collagen fibrils and other multimeric structures / Activation of Matrix Metalloproteinases / Collagen degradation / response to amyloid-beta / collagen catabolic process / negative regulation of reactive oxygen species metabolic process / extracellular matrix disassembly ...stromelysin 1 / cellular response to UV-A / regulation of neuroinflammatory response / Assembly of collagen fibrils and other multimeric structures / Activation of Matrix Metalloproteinases / Collagen degradation / response to amyloid-beta / collagen catabolic process / negative regulation of reactive oxygen species metabolic process / extracellular matrix disassembly / Degradation of the extracellular matrix / extracellular matrix organization / EGFR Transactivation by Gastrin / extracellular matrix / cellular response to nitric oxide / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / regulation of cell migration / cellular response to reactive oxygen species / cellular response to amino acid stimulus / protein catabolic process / positive regulation of protein-containing complex assembly / metalloendopeptidase activity / metallopeptidase activity / peptidase activity / cellular response to lipopolysaccharide / Interleukin-4 and Interleukin-13 signaling / endopeptidase activity / Extra-nuclear estrogen signaling / innate immune response / serine-type endopeptidase activity / mitochondrion / proteolysis / extracellular space / extracellular region / zinc ion binding / nucleus / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å | ||||||
 Authors Authors | Finzel, B.C. / Bryant Junior, G.L. / Baldwin, E.T. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 1998 Journal: Protein Sci. / Year: 1998Title: Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity. Authors: Finzel, B.C. / Baldwin, E.T. / Bryant Jr., G.L. / Hess, G.F. / Wilks, J.W. / Trepod, C.M. / Mott, J.E. / Marshall, V.P. / Petzold, G.L. / Poorman, R.A. / O'Sullivan, T.J. / Schostarez, H.J. / Mitchell, M.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
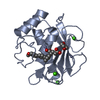
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2usn.cif.gz 2usn.cif.gz | 51.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2usn.ent.gz pdb2usn.ent.gz | 36.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2usn.json.gz 2usn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/us/2usn https://data.pdbj.org/pub/pdb/validation_reports/us/2usn ftp://data.pdbj.org/pub/pdb/validation_reports/us/2usn ftp://data.pdbj.org/pub/pdb/validation_reports/us/2usn | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1usnSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 18595.684 Da / Num. of mol.: 1 / Fragment: CATALYTIC DOMAIN RESIDUES 83 - 247 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Cell: FIBROBLAST / Species (production host): Escherichia coli / Production host: Homo sapiens (human) / Cell: FIBROBLAST / Species (production host): Escherichia coli / Production host:  | ||||||
|---|---|---|---|---|---|---|---|
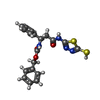
| #2: Chemical | | #3: Chemical | #4: Chemical | ChemComp-IN8 / [ | #5: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.46 Å3/Da / Density % sol: 49.95 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 7 Details: HANGING DROP VAPOR DIFFUSION., pH 7.0, vapor diffusion - hanging drop | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop / Details: inhibitor solution : protein = 1:10 | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: SIEMENS / Wavelength: 1.5418 ROTATING ANODE / Type: SIEMENS / Wavelength: 1.5418 |
| Detector | Type: SIEMENS / Detector: AREA DETECTOR / Date: Apr 29, 1996 |
| Radiation | Monochromator: GRAPHITE(002) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→20 Å / Num. obs: 8566 / % possible obs: 88 % / Observed criterion σ(I): 1 / Redundancy: 4.52 % / Rsym value: 0.086 / Net I/σ(I): 8 |
| Reflection shell | Resolution: 2.2→2.33 Å / Mean I/σ(I) obs: 4.3 / Rsym value: 0.192 / % possible all: 77 |
| Reflection | *PLUS Num. measured all: 38781 / Rmerge(I) obs: 0.086 |
| Reflection shell | *PLUS % possible obs: 77 % / Rmerge(I) obs: 0.192 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PROTEIN COMPONENT OF STROMELYSIN-1 MODEL WITH OTHER INHIBITOR (1USN) Resolution: 2.2→10 Å / σ(F): 2 Details: SEILECKI ET AL (JMB V134, 1979) PARAMETERS USED IN REFINEMENT. THROUGHOUT THE MODEL, RESIDUES ARE NUMBERED RELATIVE TO THE SEQUENCE OF THE PROENZYME. THE FIRST RESIDUE OF THE MATURE ACTIVE ...Details: SEILECKI ET AL (JMB V134, 1979) PARAMETERS USED IN REFINEMENT. THROUGHOUT THE MODEL, RESIDUES ARE NUMBERED RELATIVE TO THE SEQUENCE OF THE PROENZYME. THE FIRST RESIDUE OF THE MATURE ACTIVE CATALYTIC DOMAIN IS PHE 83. RESIDUES 190-191 ARE PARTIALY DISORDERED. THE CONFORMATION OF THESE ATOMS IS UNCERTAIN. SIDECHAINS OF RESIDUES ASP 111 AND GLU 216 ARE MODELED IN TWO DISCRETE CONFORMATIONS. THE MODEL INCLUDES A CATALYTIC ZN+2 (257), A STRUCTURAL ZN+2 (258), AND THREE BOUND CA+2 IONS (259-261). A THIRD ZN+2 HAS BEEN IDENTIFIED IN THIS CRYSTAL FORM AND RESTS ON THE CRYSTALLOGRAPHIC THREE-FOLD AXIS, COORDINATED BY THREE CRYSTALLOGRAPHICALLY RELATED OCCURANCES OF HIS 96 NE2 AND AN UNKNOWN FOURTH LIGAND. THE FOURTH LIGAND IS MODELED WITH TWO ATOMS. ONE (HOH 263 O) COORDINATED TO THE ZN 262 ALSO SITS ON THE THREE-FOLD AXIS. ONE ATOM (HOH 264 O) BONDED (2.05 ANGSTROMS) TO IT. THIS SECOND ATOM IS REPLICATED THREE TIMES BY SYMMETRY. THE FOURTH LIGAND THEREFORE RESEMBLES A CARBONATE OR NITRITE ION.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 19.11 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: PROFFT / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 2.2 Å / Lowest resolution: 2.3 Å |
 Movie
Movie Controller
Controller











 PDBj
PDBj