[English] 日本語
 Yorodumi
Yorodumi- PDB-2tbs: COLD-ADAPTION OF ENZYMES: STRUCTURAL COMPARISON BETWEEN SALMON AN... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2tbs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | COLD-ADAPTION OF ENZYMES: STRUCTURAL COMPARISON BETWEEN SALMON AND BOVINE TRYPSINS | |||||||||
 Components Components | TRYPSIN | |||||||||
 Keywords Keywords | HYDROLASE(SERINE PROTEINASE) | |||||||||
| Function / homology |  Function and homology information Function and homology informationtrypsin / digestion / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.8 Å X-RAY DIFFRACTION / Resolution: 1.8 Å | |||||||||
 Authors Authors | Smalas, A.O. | |||||||||
 Citation Citation |  Journal: Proteins / Year: 1994 Journal: Proteins / Year: 1994Title: Cold adaption of enzymes: structural comparison between salmon and bovine trypsins. Authors: Smalas, A.O. / Heimstad, E.S. / Hordvik, A. / Willassen, N.P. / Male, R. #1:  Journal: Acta Crystallogr.,Sect.D / Year: 1993 Journal: Acta Crystallogr.,Sect.D / Year: 1993Title: Crystallization and Preliminary X-Ray Crystallographic Studies of Benzamidine-Inhibited Trypsin from the North Atlantic Salmon (Salmo Salar) Authors: Smalas, A.O. / Hordvik, A. #2:  Journal: J.Mol.Biol. / Year: 1990 Journal: J.Mol.Biol. / Year: 1990Title: Crystallization and Preliminary X-Ray Crystallographic Studies of Benzamidine-Inhibited Trypsin from the North Atlantic Salmon (Salmo Salar) Authors: Smalas, A.O. / Hordvik, A. / Hansen, L.K. / Hough, E. / Jynge, K. #3:  Journal: Acta Crystallogr.,Sect.B / Year: 1983 Journal: Acta Crystallogr.,Sect.B / Year: 1983Title: The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors Authors: Marquart, M. / Walter, J. / Deisenhofer, J. / Bode, W. / Huber, R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2tbs.cif.gz 2tbs.cif.gz | 67.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2tbs.ent.gz pdb2tbs.ent.gz | 42.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2tbs.json.gz 2tbs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tb/2tbs https://data.pdbj.org/pub/pdb/validation_reports/tb/2tbs ftp://data.pdbj.org/pub/pdb/validation_reports/tb/2tbs ftp://data.pdbj.org/pub/pdb/validation_reports/tb/2tbs | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 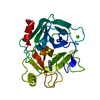
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 23784.637 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  |
|---|---|
| #2: Chemical | ChemComp-CA / |
| #3: Chemical | ChemComp-BEN / |
| #4: Water | ChemComp-HOH / |
| Has protein modification | Y |
| Nonpolymer details | THE CALCIUM IS ION IS BOUND IN A MANNER SIMILAR TO THAT OBSERVED IN BOVINE TRYPSIN. |
| Sequence details | THE AMINO ACID NUMBERING SCHEME USED IS ADOPTED FROM CHYMOTRYPS |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.15 Å3/Da / Density % sol: 42.7 % | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 28-30 ℃ / pH: 5 / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
- Processing
Processing
| Software | Name: PROLSQ / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.8→6 Å / Num. reflection obs: 14474 / σ(F): 3 Details: THE STRUCTURE WAS SOLVED BY MOLECULAR REPLACEMENT METHODS, USING THE MERLOT-PACKAGE (FITZGERALD, P. (1988) J.APPL. CRYST., 21, 273-278), AND THE REFINED MODEL OF BOVINE TRYPSIN AS SEARCH ...Details: THE STRUCTURE WAS SOLVED BY MOLECULAR REPLACEMENT METHODS, USING THE MERLOT-PACKAGE (FITZGERALD, P. (1988) J.APPL. CRYST., 21, 273-278), AND THE REFINED MODEL OF BOVINE TRYPSIN AS SEARCH MODEL (PROTEIN DATA BANK, ENTRY 3PTB). THIS ENTRY WAS REFINED USING A NON-STANDARD SETTING FOR THE SPACE GROUP P 21 21 2. THE FOLLOWING SYMMETRY OPERATORS MUST BE USED TO GENERATE CRYSTALLOGRAPHICALLY RELATED MOLECULES. X, Y, Z -X, 1/2+Y, -Z 1/2+X, -Y, -Z 1/2-X, 1/2-Y, Z | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→6 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: PROLSQ / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.165 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller




















 PDBj
PDBj







