[English] 日本語
 Yorodumi
Yorodumi- PDB-2rqw: Solution structure of Bem1p SH3CI domain complexed with Ste20p-PR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2rqw | ||||||
|---|---|---|---|---|---|---|---|
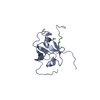
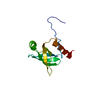
| Title | Solution structure of Bem1p SH3CI domain complexed with Ste20p-PRR peptide | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / Bem1p / Ste20p / SH3CI / PRR / Cytoplasm / Cytoskeleton / SH3 domain / Cell cycle / Pheromone response / Serine/threonine-protein kinase | ||||||
| Function / homology |  Function and homology information Function and homology informationsterol import / negative regulation of sterol import / CD28 dependent Vav1 pathway / osmosensory signaling pathway via Sho1 osmosensor / positive regulation of vacuole fusion, non-autophagic / RHO GTPases activate PAKs / bipolar cellular bud site selection / RHO GTPases Activate NADPH Oxidases / signal transduction involved in filamentous growth / : ...sterol import / negative regulation of sterol import / CD28 dependent Vav1 pathway / osmosensory signaling pathway via Sho1 osmosensor / positive regulation of vacuole fusion, non-autophagic / RHO GTPases activate PAKs / bipolar cellular bud site selection / RHO GTPases Activate NADPH Oxidases / signal transduction involved in filamentous growth / : / RHOV GTPase cycle / budding cell apical bud growth / histone H2BS14 kinase activity / conjugation with cellular fusion / cell morphogenesis involved in conjugation with cellular fusion / response to pheromone triggering conjugation with cellular fusion / site of polarized growth / Regulation of actin dynamics for phagocytic cup formation / RHOU GTPase cycle / RHOQ GTPase cycle / pseudohyphal growth / PAR polarity complex / pheromone-dependent signal transduction involved in conjugation with cellular fusion / cellular bud site selection / invasive growth in response to glucose limitation / vacuole inheritance / MAP kinase kinase kinase kinase activity / incipient cellular bud site / cellular bud tip / maintenance of protein location / regulation of exit from mitosis / cellular bud neck / regulation of Rho protein signal transduction / phosphatidylinositol-3-phosphate binding / mating projection tip / MAPK6/MAPK4 signaling / regulation of MAPK cascade / stress granule assembly / cellular response to starvation / cellular response to hydrogen peroxide / MAPK cascade / cell cortex / protein-macromolecule adaptor activity / molecular adaptor activity / cytoskeleton / protein kinase activity / non-specific serine/threonine protein kinase / intracellular signal transduction / positive regulation of apoptotic process / negative regulation of gene expression / protein serine kinase activity / protein serine/threonine kinase activity / mRNA binding / negative regulation of transcription by RNA polymerase II / mitochondrion / ATP binding / nucleus / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | SOLUTION NMR / torsion angle dynamics | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Takaku, T. / Ogura, K. / Inagaki, F. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2010 Journal: J.Biol.Chem. / Year: 2010Title: Solution structure of a novel Cdc42-binding module of Bem1 and its interaction with Ste20 and Cdc42 Authors: Takaku, T. / Ogura, K. / Kumeta, H. / Yoshida, N. / Inagaki, F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2rqw.cif.gz 2rqw.cif.gz | 801.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2rqw.ent.gz pdb2rqw.ent.gz | 679.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2rqw.json.gz 2rqw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rq/2rqw https://data.pdbj.org/pub/pdb/validation_reports/rq/2rqw ftp://data.pdbj.org/pub/pdb/validation_reports/rq/2rqw ftp://data.pdbj.org/pub/pdb/validation_reports/rq/2rqw | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 11414.939 Da / Num. of mol.: 1 / Fragment: SH3CI domain, residues 156-260 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 2331.562 Da / Num. of mol.: 1 / Fragment: BEM1-binding domain, residues 463-486 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Production host:  |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.5mM [U-99% 13C; U-99% 15N] protein; 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O |
|---|---|
| Sample | Conc.: 0.5 mM / Component: entity-1 / Isotopic labeling: [U-99% 13C; U-99% 15N] |
| Sample conditions | Ionic strength: 2 / pH: 6.5 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 800 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 1 | ||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 100 / Conformers submitted total number: 20 / Representative conformer: 1 |
 Movie
Movie Controller
Controller











 PDBj
PDBj





 CYANA
CYANA