[English] 日本語
 Yorodumi
Yorodumi- PDB-2rel: SOLUTION STRUCTURE OF R-ELAFIN, A SPECIFIC INHIBITOR OF ELASTASE,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2rel | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF R-ELAFIN, A SPECIFIC INHIBITOR OF ELASTASE, NMR, 11 STRUCTURES | |||||||||
 Components Components | R-ELAFIN | |||||||||
 Keywords Keywords | SERINE PROTEASE INHIBITOR / R-ELAFIN / ELASTASE INHIBITOR | |||||||||
| Function / homology |  Function and homology information Function and homology informationcornification / copulation / structural constituent of skin epidermis / Formation of the cornified envelope / cornified envelope / Antimicrobial peptides / endopeptidase inhibitor activity / serine-type endopeptidase inhibitor activity / extracellular matrix / antibacterial humoral response ...cornification / copulation / structural constituent of skin epidermis / Formation of the cornified envelope / cornified envelope / Antimicrobial peptides / endopeptidase inhibitor activity / serine-type endopeptidase inhibitor activity / extracellular matrix / antibacterial humoral response / innate immune response / extracellular space / extracellular region / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | SOLUTION NMR / DISTANCE GEOMETRY, SIMULATED ANNEALING | |||||||||
 Authors Authors | Francart, C. / Dauchez, M. / Alix, A.J.P. / Lippens, G. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1997 Journal: J.Mol.Biol. / Year: 1997Title: Solution structure of R-elafin, a specific inhibitor of elastase. Authors: Francart, C. / Dauchez, M. / Alix, A.J. / Lippens, G. #1:  Journal: Biochem.Biophys.Res.Commun. / Year: 1992 Journal: Biochem.Biophys.Res.Commun. / Year: 1992Title: Synthesis and Structure-Activity Relationships of Elafin, an Elastase-Specific Inhibitor Authors: Tsunemi, M. / Kato, H. / Nishiuchi, Y. / Kumagaye, S. / Sakakibara, S. #2:  Journal: J.Biol.Chem. / Year: 1991 Journal: J.Biol.Chem. / Year: 1991Title: Erratum. Elafin: An Elastase-Specific Inhibitor of Human Skin. Purification, Characterization, and Complete Amino Acid Sequence Authors: Wiedow, O. / Schroder, J.M. / Gregory, H. / Young, J.A. / Christophers, E. #3:  Journal: J.Biol.Chem. / Year: 1990 Journal: J.Biol.Chem. / Year: 1990Title: Elafin: An Elastase-Specific Inhibitor of Human Skin. Purification, Characterization, and Complete Amino Acid Sequence Authors: Wiedow, O. / Schroder, J.M. / Gregory, H. / Young, J.A. / Christophers, E. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2rel.cif.gz 2rel.cif.gz | 186.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2rel.ent.gz pdb2rel.ent.gz | 151.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2rel.json.gz 2rel.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/re/2rel https://data.pdbj.org/pub/pdb/validation_reports/re/2rel ftp://data.pdbj.org/pub/pdb/validation_reports/re/2rel ftp://data.pdbj.org/pub/pdb/validation_reports/re/2rel | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 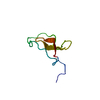
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6014.225 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) Homo sapiens (human)Description: THE POLYPEPTIDE MAY BE OBTAINED BY EXPRESSION USING PLASMIDIC EXPRESSION SYSTEMS IN HOSTS SUCH AS ESCHERICHIA COLI AND YEAST, THE POLYPEPTIDE BEING ALSO OBTAINABLE FROM PSORIATIC PLAQUES. ...Description: THE POLYPEPTIDE MAY BE OBTAINED BY EXPRESSION USING PLASMIDIC EXPRESSION SYSTEMS IN HOSTS SUCH AS ESCHERICHIA COLI AND YEAST, THE POLYPEPTIDE BEING ALSO OBTAINABLE FROM PSORIATIC PLAQUES. PATENT NUMBER 5,464,822 DATE OF PATENT NOV. 7, 1995 BY CHRISTOPHERS E., WIEDOW O., SCHRODER J.M. THE PROTEIN WAS PROVIDED BY ZENECA PHARMACEUTICALS. Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 7 / Temperature: 293 K |
|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DMX 600 / Manufacturer: Bruker / Model: DMX 600 / Field strength: 600 MHz |
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: DISTANCE GEOMETRY, SIMULATED ANNEALING / Software ordinal: 1 | ||||||||||||
| NMR ensemble | Conformer selection criteria: NOE AND DIHEDRAL ANGLE VIOLATIONS, ENERGY Conformers calculated total number: 200 / Conformers submitted total number: 11 |
 Movie
Movie Controller
Controller









 PDBj
PDBj

 HSQC
HSQC