[English] 日本語
 Yorodumi
Yorodumi- PDB-2qsz: Human nicotinamide riboside kinase 1 in complex with nicotinamide... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2qsz | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human nicotinamide riboside kinase 1 in complex with nicotinamide mononucleotide | ||||||
 Components Components | Nicotinamide riboside kinase 1 | ||||||
 Keywords Keywords | TRANSFERASE / non-protein kinase / NAD+ / nicotinamide riboside / nrk1 / nicotinamide riboside kinase activity / nicotinic acid riboside kinase activity / NAD biosynthesis / ADP / STRUCTURAL GENOMICS / STRUCTURAL GENOMICS CONSORTIUM / SGC / Alternative splicing / ATP-binding / Nucleotide-binding / Pyridine nucleotide biosynthesis | ||||||
| Function / homology |  Function and homology information Function and homology informationnicotinate riboside kinase / : / nicotinate riboside kinase activity / ribosylnicotinamide kinase / ribosylnicotinamide kinase activity / Nicotinate metabolism / NAD+ biosynthetic process / NAD+ metabolic process / ATP binding / metal ion binding ...nicotinate riboside kinase / : / nicotinate riboside kinase activity / ribosylnicotinamide kinase / ribosylnicotinamide kinase activity / Nicotinate metabolism / NAD+ biosynthetic process / NAD+ metabolic process / ATP binding / metal ion binding / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.9 Å molecular replacement / Resolution: 1.9 Å | ||||||
 Authors Authors | Rabeh, W.M. / Tempel, W. / Nedyalkova, L. / Landry, R. / Arrowsmith, C.H. / Edwards, A.M. / Sundstrom, M. / Weigelt, J. / Bochkarev, A. / Brenner, C. ...Rabeh, W.M. / Tempel, W. / Nedyalkova, L. / Landry, R. / Arrowsmith, C.H. / Edwards, A.M. / Sundstrom, M. / Weigelt, J. / Bochkarev, A. / Brenner, C. / Park, H. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: Plos Biol. / Year: 2007 Journal: Plos Biol. / Year: 2007Title: Nicotinamide Riboside Kinase Structures Reveal New Pathways to NAD(+). Authors: Tempel, W. / Rabeh, W.M. / Bogan, K.L. / Belenky, P. / Wojcik, M. / Seidle, H.F. / Nedyalkova, L. / Yang, T. / Sauve, A.A. / Park, H.W. / Brenner, C. | ||||||
| History |
| ||||||
| Remark 300 | BIOMOLECULE: 1 SEE REMARK 350 FOR THE PROGRAM GENERATED ASSEMBLY INFORMATION FOR THE STRUCTURE IN ... BIOMOLECULE: 1 SEE REMARK 350 FOR THE PROGRAM GENERATED ASSEMBLY INFORMATION FOR THE STRUCTURE IN THIS ENTRY. AUTHORS STATE THAT THE BIOLOGICAL UNIT OF THIS POLYPEPTIDE IS UNKNOWN. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2qsz.cif.gz 2qsz.cif.gz | 59 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2qsz.ent.gz pdb2qsz.ent.gz | 41 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2qsz.json.gz 2qsz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qs/2qsz https://data.pdbj.org/pub/pdb/validation_reports/qs/2qsz ftp://data.pdbj.org/pub/pdb/validation_reports/qs/2qsz ftp://data.pdbj.org/pub/pdb/validation_reports/qs/2qsz | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2p0eC  2qsySC  2qt0C  2qt1C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | not known |
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 24373.371 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: NRK1, C9orf95 / Plasmid: p28a-LIC / Production host: Homo sapiens (human) / Gene: NRK1, C9orf95 / Plasmid: p28a-LIC / Production host:  References: UniProt: Q9NWW6, Transferases; Transferring phosphorus-containing groups; Phosphotransferases with an alcohol group as acceptor |
|---|
-Non-polymers , 5 types, 113 molecules 

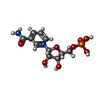






| #2: Chemical | ChemComp-PO4 / | ||
|---|---|---|---|
| #3: Chemical | ChemComp-CL / | ||
| #4: Chemical | ChemComp-NMN / | ||
| #5: Chemical | ChemComp-UNX / #6: Water | ChemComp-HOH / | |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.55 Å3/Da / Density % sol: 51.74 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 20% PEG 3350, 0.2M Sodium phosphate monobasic, 0.15M D(+)sucrose, 0.1M HEPES pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU FR-E+ SUPERBRIGHT / Wavelength: 1.5418 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RIGAKU RAXIS / Detector: IMAGE PLATE / Date: Jan 26, 2007 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.9→30 Å / Num. obs: 19738 / % possible obs: 99.1 % / Redundancy: 4.8 % / Rmerge(I) obs: 0.138 / Χ2: 1.911 / Net I/σ(I): 6.4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 2QSY Resolution: 1.9→23.64 Å / Cor.coef. Fo:Fc: 0.942 / Cor.coef. Fo:Fc free: 0.927 / WRfactor Rfree: 0.218 / WRfactor Rwork: 0.188 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.16 / ESU R Free: 0.149 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS. Prodrg, arp/warp, coot, molprobity programs have also been used in refinement
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.061 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→23.64 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj



