+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2pik | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CALICHEAMICIN GAMMA1I-DNA COMPLEX, NMR, 6 STRUCTURES | |||||||||||||||||||||
 Components Components | DNA(5'-D(C* Keywords KeywordsDNA / DEOXYRIBONUCLEIC ACID / CALICHEAMICIN GAMMA1I - DNA COMPLEX / ENEDIYNE ALIGNMENT IN MINOR GROOVE / SACCHARIDE DNA MINOR GROOVE INTERACTIONS / INTERMOLECULAR DRUG IODINE-GUANINE AMINO PROTON INTERACTIONS / SPECIFICITY AND CLEAVAGE PROCESS | Function / homology | 2,6-dideoxy-4-thio-beta-D-allopyranose / Chem-HIB / 3-O-methyl-alpha-L-rhamnopyranose / Chem-MTC / DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR / simulated annealing |  Authors AuthorsKumar, R.A. / Ikemoto, N. / Patel, D.J. |  Citation Citation Journal: J.Mol.Biol. / Year: 1997 Journal: J.Mol.Biol. / Year: 1997Title: Solution structure of the calicheamicin gamma 1I-DNA complex. Authors: Kumar, R.A. / Ikemoto, N. / Patel, D.J. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1995 Journal: Proc.Natl.Acad.Sci.USA / Year: 1995Title: Calicheamicin-DNA Complexes: Warhead Alignment and Saccharide Recognition of the Minor Groove Authors: Ikemoto, N. / Kumar, R.A. / Ling, T.T. / Ellestad, G.A. / Danishefsky, S.J. / Patel, D.J. History |
|
- Structure visualization
Structure visualization
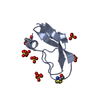
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2pik.cif.gz 2pik.cif.gz | 99.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2pik.ent.gz pdb2pik.ent.gz | 76 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2pik.json.gz 2pik.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pi/2pik https://data.pdbj.org/pub/pdb/validation_reports/pi/2pik ftp://data.pdbj.org/pub/pdb/validation_reports/pi/2pik ftp://data.pdbj.org/pub/pdb/validation_reports/pi/2pik | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
-DNA chain , 1 types, 1 molecules A
| #1: DNA chain | Mass: 7038.531 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|
-Sugars , 3 types, 3 molecules 

| #2: Polysaccharide | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D- ...2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose Type: oligosaccharide / Mass: 320.382 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #3: Sugar | ChemComp-DSR / |
| #5: Sugar | ChemComp-MRP / |
-Non-polymers , 2 types, 2 molecules 
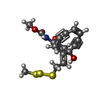
| #4: Chemical | ChemComp-HIB / |
|---|---|
| #6: Chemical | ChemComp-MTC / [ |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Sample conditions |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: simulated annealing / Software ordinal: 1 | ||||||||||||
| NMR ensemble | Conformers calculated total number: 6 / Conformers submitted total number: 6 |
 Movie
Movie Controller
Controller









 PDBj
PDBj










































 X-PLOR
X-PLOR