+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2n87 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
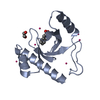
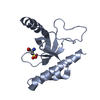
| Title | Solution structure of the PPIase domain of TbPar42 | |||||||||
 Components Components | Uncharacterized protein | |||||||||
 Keywords Keywords | ISOMERASE / PPIase domain / Parvulin | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / mRNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | SOLUTION NMR / torsion angle dynamics, simulated annealing, molecular dynamics | |||||||||
| Model details | lowest energy, model1 | |||||||||
 Authors Authors | Rehic, E. / Bayer, P. | |||||||||
 Citation Citation |  Journal: Biomolecules / Year: 2019 Journal: Biomolecules / Year: 2019Title: Structural Analysis of the 42 kDa Parvulin of Trypanosoma brucei. Authors: Rehic, E. / Hoenig, D. / Kamba, B.E. / Goehring, A. / Hofmann, E. / Gasper, R. / Matena, A. / Bayer, P. #1: Journal: Febs Lett. / Year: 2010 Title: Functional characterization of two novel parvulins in Trypanosoma brucei. Authors: Goh, J.Y. / Lai, C.Y. / Tan, L.C. / Yang, D. / He, C.Y. / Liou, Y.C. #2: Journal: Biochim.Biophys.Acta / Year: 2010 Title: Identification of an atypical peptidyl-prolyl cis/trans isomerase from trypanosomatids. Authors: Erben, E.D. / Valguarnera, E. / Nardelli, S. / Chung, J. / Daum, S. / Potenza, M. / Schenkman, S. / Tellez-Inon, M.T. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2n87.cif.gz 2n87.cif.gz | 431.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2n87.ent.gz pdb2n87.ent.gz | 366.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2n87.json.gz 2n87.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2n87_validation.pdf.gz 2n87_validation.pdf.gz | 398.3 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2n87_full_validation.pdf.gz 2n87_full_validation.pdf.gz | 466.1 KB | Display | |
| Data in XML |  2n87_validation.xml.gz 2n87_validation.xml.gz | 21.8 KB | Display | |
| Data in CIF |  2n87_validation.cif.gz 2n87_validation.cif.gz | 33.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/n8/2n87 https://data.pdbj.org/pub/pdb/validation_reports/n8/2n87 ftp://data.pdbj.org/pub/pdb/validation_reports/n8/2n87 ftp://data.pdbj.org/pub/pdb/validation_reports/n8/2n87 | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 13522.224 Da / Num. of mol.: 1 / Fragment: residues 264-383 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: 927/4 GUTat10.1 / Gene: Tb927.7.2480 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sample conditions | pH: 6.26 / Temperature: 300 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance Ultrashield / Manufacturer: Bruker / Model: Avance Ultrashield / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics, simulated annealing, molecular dynamics Software ordinal: 1 | ||||||||||||||||||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 2268 / NOE intraresidue total count: 455 / NOE long range total count: 802 / NOE medium range total count: 443 / NOE sequential total count: 568 / Hydrogen bond constraints total count: 185 / Protein phi angle constraints total count: 103 / Protein psi angle constraints total count: 103 | ||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 60 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller













 PDBj
PDBj



 HSQC
HSQC