| Entry | Database: PDB / ID: 2n2n
|
|---|
| Title | Tom1 negatively modulates binding of Tollip to phosphatidylinositol 3-phosphate via a coupled folding and binding mechanism |
|---|
 Components Components | Target of Myb protein 1 |
|---|
 Keywords Keywords | PROTEIN TRANSPORT |
|---|
| Function / homology |  Function and homology information Function and homology information
myosin VI binding / substrate localization to autophagosome / regulation of endosome organization / clathrin heavy chain binding / phosphatidylinositol-5-phosphate binding / autophagosome-lysosome fusion / positive regulation of autophagosome maturation / endosomal transport / azurophil granule membrane / clathrin binding ...myosin VI binding / substrate localization to autophagosome / regulation of endosome organization / clathrin heavy chain binding / phosphatidylinositol-5-phosphate binding / autophagosome-lysosome fusion / positive regulation of autophagosome maturation / endosomal transport / azurophil granule membrane / clathrin binding / polyubiquitin modification-dependent protein binding / specific granule membrane / ubiquitin binding / endocytosis / protein transport / early endosome membrane / early endosome / endosome / endosome membrane / Neutrophil degranulation / Golgi apparatus / signal transduction / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosolSimilarity search - Function Target of Myb protein 1 / Methane Monooxygenase Hydroxylase; Chain G, domain 1 - #160 / GAT domain / GAT domain superfamily / GAT domain / GAT domain profile. / VHS domain / VHS domain / VHS domain profile. / Domain present in VPS-27, Hrs and STAM ...Target of Myb protein 1 / Methane Monooxygenase Hydroxylase; Chain G, domain 1 - #160 / GAT domain / GAT domain superfamily / GAT domain / GAT domain profile. / VHS domain / VHS domain / VHS domain profile. / Domain present in VPS-27, Hrs and STAM / ENTH/VHS / Methane Monooxygenase Hydroxylase; Chain G, domain 1 / Up-down Bundle / Mainly AlphaSimilarity search - Domain/homology |
|---|
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
|---|
| Method | SOLUTION NMR / DISTANCE GEOMETRY, SIMULATED ANNEALING |
|---|
| Model details | score result from Rosetta, model1 |
|---|
 Authors Authors | Xiao, S. / Armstrong, G. / Capelluto, D. |
|---|
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015
Title: Tom1 Modulates Binding of Tollip to Phosphatidylinositol 3-Phosphate via a Coupled Folding and Binding Mechanism.
Authors: Xiao, S. / Brannon, M.K. / Zhao, X. / Fread, K.I. / Ellena, J.F. / Bushweller, J.H. / Finkielstein, C.V. / Armstrong, G.S. / Capelluto, D.G. |
|---|
| History | | Deposition | May 11, 2015 | Deposition site: BMRB / Processing site: RCSB |
|---|
| Revision 1.0 | Sep 16, 2015 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Oct 21, 2015 | Group: Database references |
|---|
| Revision 1.2 | Jun 14, 2023 | Group: Data collection / Database references / Other
Category: database_2 / pdbx_database_status ...database_2 / pdbx_database_status / pdbx_nmr_software / pdbx_nmr_spectrometer / struct_ref_seq_dif
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession ..._database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_nmr_data / _pdbx_nmr_software.name / _pdbx_nmr_spectrometer.model / _struct_ref_seq_dif.details |
|---|
| Revision 1.3 | May 15, 2024 | Group: Data collection / Database references / Category: chem_comp_atom / chem_comp_bond / database_2 / Item: _database_2.pdbx_DOI |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) Authors
Authors Citation
Citation Journal: Structure / Year: 2015
Journal: Structure / Year: 2015 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2n2n.cif.gz
2n2n.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2n2n.ent.gz
pdb2n2n.ent.gz PDB format
PDB format 2n2n.json.gz
2n2n.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/n2/2n2n
https://data.pdbj.org/pub/pdb/validation_reports/n2/2n2n ftp://data.pdbj.org/pub/pdb/validation_reports/n2/2n2n
ftp://data.pdbj.org/pub/pdb/validation_reports/n2/2n2n Links
Links Assembly
Assembly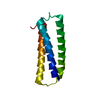
 Components
Components Homo sapiens (human) / Gene: TOM1 / Production host:
Homo sapiens (human) / Gene: TOM1 / Production host: 
 Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller









 PDBj
PDBj
 HSQC
HSQC