[English] 日本語
 Yorodumi
Yorodumi- PDB-2mj2: Structure of the dimerization domain of the human polyoma, JC vir... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mj2 | ||||||
|---|---|---|---|---|---|---|---|
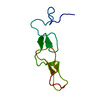
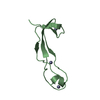
| Title | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | ||||||
 Components Components | Agnoprotein | ||||||
 Keywords Keywords | VIRAL PROTEIN / Agnoprotein / polyomavirus JCV / DNA replication / progressive multifocal leukoencephalopathy | ||||||
| Function / homology |  Function and homology information Function and homology informationhost cell rough endoplasmic reticulum membrane / host cell nuclear membrane / channel activity / monoatomic ion transmembrane transport / host cell plasma membrane / DNA binding / membrane Similarity search - Function | ||||||
| Biological species |   JC polyomavirus JC polyomavirus | ||||||
| Method | SOLUTION NMR / DGSA-distance geometry simulated annealing | ||||||
| Model details | lowest energy, model 1 | ||||||
 Authors Authors | Coric, P. / Saribas, S.A. / Abou-Gharbia, M. / Childers, W. / White, M. / Bouaziz, S. / Safak, M. | ||||||
 Citation Citation |  Journal: J.Virol. / Year: 2014 Journal: J.Virol. / Year: 2014Title: The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix Authors: Coric, P. / Saribas, S.A. / Abou-Gharbia, M. / Childers, W. / White, M. / Bouaziz, S. / Safak, M. #1: Journal: Virology / Year: 2013 Title: Essential roles of Leu/Ile/Phe-rich domain of JC virus agnoprotein in dimer/oligomer formation, protein stability and splicing of viral transcripts. Authors: Sami Saribas, A. / Abou-Gharbia, M. / Childers, W. / Sariyer, I.K. / White, M.K. / Safak, M. #2: Journal: Virology / Year: 2011 Title: Human polyomavirus JC small regulatory agnoprotein forms highly stable dimers and oligomers: implications for their roles in agnoprotein function. Authors: Saribas, A.S. / Arachea, B.T. / White, M.K. / Viola, R.E. / Safak, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mj2.cif.gz 2mj2.cif.gz | 202.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mj2.ent.gz pdb2mj2.ent.gz | 167.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mj2.json.gz 2mj2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2mj2_validation.pdf.gz 2mj2_validation.pdf.gz | 433.1 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2mj2_full_validation.pdf.gz 2mj2_full_validation.pdf.gz | 562.4 KB | Display | |
| Data in XML |  2mj2_validation.xml.gz 2mj2_validation.xml.gz | 22.7 KB | Display | |
| Data in CIF |  2mj2_validation.cif.gz 2mj2_validation.cif.gz | 25.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mj/2mj2 https://data.pdbj.org/pub/pdb/validation_reports/mj/2mj2 ftp://data.pdbj.org/pub/pdb/validation_reports/mj/2mj2 ftp://data.pdbj.org/pub/pdb/validation_reports/mj/2mj2 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 4207.873 Da / Num. of mol.: 1 / Fragment: UNP residues 17-52 / Source method: obtained synthetically / Source: (synth.)   JC polyomavirus / References: UniProt: P03086 JC polyomavirus / References: UniProt: P03086 |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.5 mM AGNO, 70 v/v H2O, 30 v/v [U-100% 2H] TFE, water with 30% (v/v) TFE (Trifluoroethanol, CF3CH2OH) Solvent system: water with 30% (v/v) TFE (Trifluoroethanol, CF3CH2OH) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||
| Sample conditions |
|
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: DGSA-distance geometry simulated annealing / Software ordinal: 1 | ||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 200 / Conformers submitted total number: 17 |
 Movie
Movie Controller
Controller








 PDBj
PDBj