+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2kyq | ||||||
|---|---|---|---|---|---|---|---|
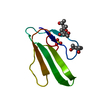
| Title | 1H, 15N, 13C chemical shifts and structure of CKR-brazzein | ||||||
 Components Components | Defensin-like protein | ||||||
 Keywords Keywords | PLANT PROTEIN / SWEET-TASTING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationdefense response to fungus / killing of cells of another organism / defense response to bacterium / extracellular region Similarity search - Function | ||||||
| Biological species |  Pentadiplandra brazzeana (plant) Pentadiplandra brazzeana (plant) | ||||||
| Method | SOLUTION NMR / torsion angle dynamics | ||||||
| Model details | closest to the average, model 9 | ||||||
 Authors Authors | Dittli, S.M. / Assadi-Porter, F.M. / Rao, H. / Tonelli, M. | ||||||
 Citation Citation |  Journal: Chem Senses / Year: 2011 Journal: Chem Senses / Year: 2011Title: Structural role of the terminal disulfide bond in the sweetness of brazzein. Authors: Dittli, S.M. / Rao, H. / Tonelli, M. / Quijada, J. / Markley, J.L. / Max, M. / Assadi-Porter, F. #1:  Journal: To be Published Journal: To be PublishedTitle: Insights into the sweetness of brazzein from beta-hairpin peptides derived from the N- and C- termini of brazzein Authors: Dittli, S.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2kyq.cif.gz 2kyq.cif.gz | 380.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2kyq.ent.gz pdb2kyq.ent.gz | 321.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2kyq.json.gz 2kyq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ky/2kyq https://data.pdbj.org/pub/pdb/validation_reports/ky/2kyq ftp://data.pdbj.org/pub/pdb/validation_reports/ky/2kyq ftp://data.pdbj.org/pub/pdb/validation_reports/ky/2kyq | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6408.243 Da / Num. of mol.: 1 / Mutation: K3C, C4K, K5R Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Pentadiplandra brazzeana (plant) / Production host: Pentadiplandra brazzeana (plant) / Production host:  |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR / Details: Triple brazzein mutant: K3C,C4K,K5R | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||
| Sample conditions | pH: 5.2 / Pressure: ambient / Temperature: 310 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 1 | ||||||||||||||||||||||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 741 / NOE intraresidue total count: 157 / NOE long range total count: 213 / NOE medium range total count: 134 / NOE sequential total count: 249 / Hydrogen bond constraints total count: 28 / Protein chi angle constraints total count: 0 / Protein other angle constraints total count: 0 / Protein phi angle constraints total count: 94 / Protein psi angle constraints total count: 94 | ||||||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller














 PDBj
PDBj

 HSQC
HSQC