[English] 日本語
 Yorodumi
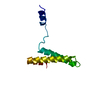
Yorodumi- PDB-2fkx: Ribosomal protein s15 from thermus thermophilus, nmr recalculated... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2fkx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Ribosomal protein s15 from thermus thermophilus, nmr recalculated structure | ||||||
 Components Components | 30S ribosomal protein S15 | ||||||
 Keywords Keywords | STRUCTURAL PROTEIN / RIBOSOMAL PROTEIN / RNA-BINDING PROTEIN / RRNA-BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationcytosolic small ribosomal subunit / rRNA binding / structural constituent of ribosome / translation Similarity search - Function | ||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | ||||||
| Method | SOLUTION NMR / SIMULATED ANNEALING, TORSION ANGLE DYNAMICS | ||||||
 Authors Authors | Malliavin, T.E. | ||||||
 Citation Citation |  Journal: Biophys.J. / Year: 2007 Journal: Biophys.J. / Year: 2007Title: The Conformational Landscape of the Ribosomal Protein S15 and Its Influence on the Protein Interaction with 16S RNA. Authors: Crety, T. / Malliavin, T.E. #1:  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: Solution structure of the ribosomal rna binding protein s15 from thermus thermophilus Authors: Berglund, H. / Rak, A. / Serganov, A. / Garber, M. / Hard, T. | ||||||
| History |
| ||||||
| Remark 950 | THIS ENTRY 2FKX REFLECTS AN ALTERNATIVE MODELING OF THE STRUCTURAL DATA IN 1AB3.MR ORIGINAL DATA ...THIS ENTRY 2FKX REFLECTS AN ALTERNATIVE MODELING OF THE STRUCTURAL DATA IN 1AB3.MR ORIGINAL DATA DETERMINED BY AUTHOR: H.BERGLUND, A.RAK, A.SERGANOV, M.GARBER, T.HARD |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2fkx.cif.gz 2fkx.cif.gz | 524 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2fkx.ent.gz pdb2fkx.ent.gz | 440.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2fkx.json.gz 2fkx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2fkx_validation.pdf.gz 2fkx_validation.pdf.gz | 357.5 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2fkx_full_validation.pdf.gz 2fkx_full_validation.pdf.gz | 463.4 KB | Display | |
| Data in XML |  2fkx_validation.xml.gz 2fkx_validation.xml.gz | 26.9 KB | Display | |
| Data in CIF |  2fkx_validation.cif.gz 2fkx_validation.cif.gz | 45.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fk/2fkx https://data.pdbj.org/pub/pdb/validation_reports/fk/2fkx ftp://data.pdbj.org/pub/pdb/validation_reports/fk/2fkx ftp://data.pdbj.org/pub/pdb/validation_reports/fk/2fkx | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 10447.213 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Thermus thermophilus (bacteria) / Gene: rpsO, rps15 / Plasmid: PET11C / Species (production host): Escherichia coli / Production host: Thermus thermophilus (bacteria) / Gene: rpsO, rps15 / Plasmid: PET11C / Species (production host): Escherichia coli / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR details | Text: AUTHOR USED THE MR DATA FROM ENTRY 1AB3. |
- Sample preparation
Sample preparation
| Sample conditions | pH: 5 / Pressure: ambient / Temperature: 303 K |
|---|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian UNITY / Manufacturer: Varian / Model: UNITY / Field strength: 500 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: SIMULATED ANNEALING, TORSION ANGLE DYNAMICS / Software ordinal: 1 Details: SIMULATED ANNEALING IN TORSION ANGLE SPACE AND IN CARTESIAN COORDINATES. | ||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 18 |
 Movie
Movie Controller
Controller











 PDBj
PDBj



