[English] 日本語
 Yorodumi
Yorodumi- PDB-2df6: Crystal Structure of the SH3 Domain of betaPIX in Complex with a ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2df6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of the SH3 Domain of betaPIX in Complex with a High Affinity Peptide from PAK2 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / SH3 domain / Peptide interaction | ||||||
| Function / homology |  Function and homology information Function and homology informationCD28 dependent Vav1 pathway / VEGFR2 mediated vascular permeability / RHO GTPases activate PAKs / Sema3A PAK dependent Axon repulsion / CD209 (DC-SIGN) signaling / VEGFA-VEGFR2 Pathway / negative regulation of microtubule nucleation / Generation of second messenger molecules / Ephrin signaling / presynaptic actin cytoskeleton organization ...CD28 dependent Vav1 pathway / VEGFR2 mediated vascular permeability / RHO GTPases activate PAKs / Sema3A PAK dependent Axon repulsion / CD209 (DC-SIGN) signaling / VEGFA-VEGFR2 Pathway / negative regulation of microtubule nucleation / Generation of second messenger molecules / Ephrin signaling / presynaptic actin cytoskeleton organization / EGFR downregulation / RHOU GTPase cycle / RHOV GTPase cycle / RAC2 GTPase cycle / RHOH GTPase cycle / NRAGE signals death through JNK / Smooth Muscle Contraction / G alpha (12/13) signalling events / MAPK6/MAPK4 signaling / RHOG GTPase cycle / RHOQ GTPase cycle / RAC1 GTPase cycle / postsynaptic actin cytoskeleton organization / RHOA GTPase cycle / protein localization to cell-cell junction / storage vacuole / astrocyte cell migration / FCERI mediated MAPK activation / positive regulation of growth hormone secretion / dendritic spine development / bicellular tight junction assembly / negative regulation of stress fiber assembly / cardiac muscle hypertrophy / gamma-tubulin binding / adherens junction assembly / positive regulation of extrinsic apoptotic signaling pathway / lamellipodium assembly / small GTPase-mediated signal transduction / regulation of axonogenesis / mitotic spindle pole / regulation of cytoskeleton organization / Golgi organization / regulation of MAPK cascade / protein tyrosine kinase activator activity / cellular response to transforming growth factor beta stimulus / hematopoietic progenitor cell differentiation / Rho protein signal transduction / ruffle / guanyl-nucleotide exchange factor activity / secretory granule / cellular response to starvation / small GTPase binding / GABA-ergic synapse / cell-cell junction / cell migration / lamellipodium / growth cone / cell cortex / protein kinase activity / non-specific serine/threonine protein kinase / neuron projection / postsynapse / intracellular signal transduction / postsynaptic density / nuclear speck / positive regulation of apoptotic process / protein serine kinase activity / focal adhesion / protein serine/threonine kinase activity / neuronal cell body / centrosome / protein kinase binding / negative regulation of apoptotic process / perinuclear region of cytoplasm / glutamatergic synapse / protein-containing complex / ATP binding / identical protein binding / membrane / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.3 Å MOLECULAR REPLACEMENT / Resolution: 1.3 Å | ||||||
 Authors Authors | Hoelz, A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2006 Journal: J.Mol.Biol. / Year: 2006Title: Crystal Structure of the SH3 Domain of betaPIX in Complex with a High Affinity Peptide from PAK2 Authors: Hoelz, A. / Janz, J.M. / Lawrie, S.D. / Corwin, B. / Lee, A. / Sakmar, T.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2df6.cif.gz 2df6.cif.gz | 43.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2df6.ent.gz pdb2df6.ent.gz | 31.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2df6.json.gz 2df6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/df/2df6 https://data.pdbj.org/pub/pdb/validation_reports/df/2df6 ftp://data.pdbj.org/pub/pdb/validation_reports/df/2df6 ftp://data.pdbj.org/pub/pdb/validation_reports/df/2df6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2g6fSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 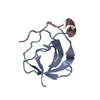
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6731.333 Da / Num. of mol.: 2 / Fragment: SH3 domain(residues 10-63) Source method: isolated from a genetically manipulated source Source: (gene. exp.)   #2: Protein/peptide | Mass: 2053.364 Da / Num. of mol.: 2 / Fragment: residues 180-197 / Source method: obtained synthetically Details: The peptide was chemically synthesized. The sequence of the peptide is naturally found in human, rat. References: GenBank: 5138914, UniProt: Q64303*PLUS, EC: 2.7.1.37 #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.08 Å3/Da / Density % sol: 40.77 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 6.7 Details: 100mM MES, 35% PEG 5000MME, 200mM ammonium sulfate, pH 6.7, VAPOR DIFFUSION, HANGING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X9A / Wavelength: 0.979191 Å / Beamline: X9A / Wavelength: 0.979191 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Sep 7, 2001 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.979191 Å / Relative weight: 1 |
| Reflection | Resolution: 1.3→30 Å / Num. all: 35167 / Num. obs: 35167 / % possible obs: 99.7 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 |
| Reflection shell | Resolution: 1.3→1.32 Å / % possible all: 99.7 |
- Processing
Processing
| Software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2G6F Resolution: 1.3→30 Å / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.3→30 Å
|
 Movie
Movie Controller
Controller












 PDBj
PDBj


