Entry Database : PDB / ID : 2chmTitle Crystal structure of N2 substituted pyrazolo pyrimidinones - a flipped binding mode in PDE5 CGMP-SPECIFIC 3', 5'-CYCLIC PHOSPHODIESTERASE, CAMP-SPECIFIC 3', 5'-CYCLIC PHOSPHODIESTERASE 4B Keywords / / / / / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species HOMO SAPIENS (human)Method / / Resolution : 1.6 Å Authors Allerton, C.M.N. / Barber, C.G. / Beaumont, K.C. / Brown, D.G. / Cole, S.M. / Ellis, D. / Lane, C.A.L. / Maw, G.N. / Mount, N.M. / Rawson, D.J. ...Allerton, C.M.N. / Barber, C.G. / Beaumont, K.C. / Brown, D.G. / Cole, S.M. / Ellis, D. / Lane, C.A.L. / Maw, G.N. / Mount, N.M. / Rawson, D.J. / Robinson, C.M. / Street, S.D.A. / Summerhill, N.W. Journal : J.Med.Chem. / Year : 2006Title : A Novel Series of Potent and Selective Pde5 Inhibitors with Potential for High and Dose-Independent Oral BioavailabilityAuthors : Allerton, C.M.N. / Barber, C.G. / Beaumont, K.C. / Brown, D.G. / Cole, S.M. / Ellis, D. / Lane, C.A.L. / Maw, G.N. / Mount, N.M. / Rawson, D.J. / Robinson, C.M. / Street, S.D.A. / Summerhill, N.W. History Deposition Mar 15, 2006 Deposition site / Processing site Revision 1.0 Jun 8, 2006 Provider / Type Revision 1.1 May 8, 2011 Group Revision 1.2 Jul 13, 2011 Group Revision 1.3 Mar 27, 2019 Group / Other / Source and taxonomyCategory entity_src_gen / pdbx_database_proc ... entity_src_gen / pdbx_database_proc / pdbx_database_status / struct_biol Item / _entity_src_gen.pdbx_host_org_strain / _pdbx_database_status.recvd_author_approvalRevision 1.4 May 1, 2024 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å
MOLECULAR REPLACEMENT / Resolution: 1.6 Å  Authors
Authors Citation
Citation Journal: J.Med.Chem. / Year: 2006
Journal: J.Med.Chem. / Year: 2006 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2chm.cif.gz
2chm.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2chm.ent.gz
pdb2chm.ent.gz PDB format
PDB format 2chm.json.gz
2chm.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ch/2chm
https://data.pdbj.org/pub/pdb/validation_reports/ch/2chm ftp://data.pdbj.org/pub/pdb/validation_reports/ch/2chm
ftp://data.pdbj.org/pub/pdb/validation_reports/ch/2chm Links
Links Assembly
Assembly
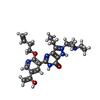
 Components
Components HOMO SAPIENS (human) / Plasmid: FASTBAC / Cell line (production host): High Five / Production host:
HOMO SAPIENS (human) / Plasmid: FASTBAC / Cell line (production host): High Five / Production host:  TRICHOPLUSIA NI (cabbage looper)
TRICHOPLUSIA NI (cabbage looper)







 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418
ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










































 PDBj
PDBj








