[English] 日本語
 Yorodumi
Yorodumi- PDB-2cd6: Refinement of RNase P P4 stemloop structure using residual dipola... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2cd6 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Refinement of RNase P P4 stemloop structure using residual dipolar coupling data, C70U mutant cobalt(III) hexammine complex | ||||||||||||||||||||
 Components Components | 5'-R(* Keywords KeywordsNUCLEIC ACID / C70U MUTANT / COBALT (III HEXAMMINE COMPLEX / METAL BINDING SITE / METAL COMPLEX / P4 STEM / RIBONUCLEASE P / RIBONUCLEIC ACID / RIBOZYME / TRANSFER RNA PROCESSING | Function / homology | COBALT HEXAMMINE(III) / RNA / RNA (> 10) |  Function and homology information Function and homology informationBiological species |  Method | SOLUTION NMR / RESTRAINED MOLECULAR DYNAMICS | Model type details | MINIMIZED AVERAGE |  Authors AuthorsSchmitz, M. | History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2cd6.cif.gz 2cd6.cif.gz | 26.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2cd6.ent.gz pdb2cd6.ent.gz | 17.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2cd6.json.gz 2cd6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  2cd6_validation.pdf.gz 2cd6_validation.pdf.gz | 326.7 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  2cd6_full_validation.pdf.gz 2cd6_full_validation.pdf.gz | 329.8 KB | Display | |
| Data in XML |  2cd6_validation.xml.gz 2cd6_validation.xml.gz | 2 KB | Display | |
| Data in CIF |  2cd6_validation.cif.gz 2cd6_validation.cif.gz | 2.4 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cd/2cd6 https://data.pdbj.org/pub/pdb/validation_reports/cd/2cd6 ftp://data.pdbj.org/pub/pdb/validation_reports/cd/2cd6 ftp://data.pdbj.org/pub/pdb/validation_reports/cd/2cd6 | HTTPS FTP |
-Related structure data
| Related structure data |  1f6xC  1f6zC  1f78C  1f79C  1f7fC  1f7gC  1f7hC  1f7iC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 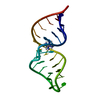
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8634.127 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: MUTATION CYTOSINE 70 URACIL / Source: (synth.)  |
|---|---|
| #2: Chemical | ChemComp-NCO / |
| Sequence details | THE SEQUENCE CORRESPONDS TO NUCLEOTIDES 66 TO 73, AND 354 TO 360 OF THE E. COLI RNASE P RNA, WITH ...THE SEQUENCE CORRESPOND |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Type: 1H-15N HSCQ |
| NMR details | Text: THE STRUCTURE WAS DETERMINED USING CONSTRAINTS FROM HOMONUCLEAR NMR EXPERIMENTS AS DESCRIBED IN ENTRY 17F9, AND ADDITIONAL RESIDUAL DIPOLAR COUPLINGS FOR IMINO RESONANCES AQCUIRED WITH AND ...Text: THE STRUCTURE WAS DETERMINED USING CONSTRAINTS FROM HOMONUCLEAR NMR EXPERIMENTS AS DESCRIBED IN ENTRY 17F9, AND ADDITIONAL RESIDUAL DIPOLAR COUPLINGS FOR IMINO RESONANCES AQCUIRED WITH AND WITHOUT 30 MG PER ML PF1 PHAGE PRESENT IN SOLUTION |
- Sample preparation
Sample preparation
| Details | Contents: 90% WATER/10% D2O |
|---|---|
| Sample conditions | Ionic strength: 100 mM / pH: 6.5 / Pressure: 1.0 atm / Temperature: 288.0 K |
-NMR measurement
| NMR spectrometer | Type: Bruker DMX / Manufacturer: Bruker / Model: DMX / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: RESTRAINED MOLECULAR DYNAMICS / Software ordinal: 1 Details: DETAILS OF THE REFINEMENT PROCEDURE AGAINST RESIDUAL DIPOLAR COUPLINGS ARE GIVEN IN THE PRIMARY CITATION ABOVE | ||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 50 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller




 ref.
ref.





 PDBj
PDBj































 Xplor-NIH
Xplor-NIH