[English] 日本語
 Yorodumi
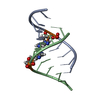
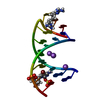
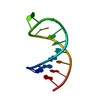
Yorodumi- PDB-229d: DNA ANALOG OF YEAST TRANSFER RNA PHE ANTICODON DOMAIN WITH MODIFI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 229d | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | DNA ANALOG OF YEAST TRANSFER RNA PHE ANTICODON DOMAIN WITH MODIFIED BASES 5-METHYL CYTOSINE AND 1-METHYL GUANINE | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / TRANSFER RNA / ANTICODON / HAIRPIN LOOP | Function / homology | DNA / DNA (> 10) |  Function and homology information Function and homology informationMethod | SOLUTION NMR |  Authors AuthorsBasti, M.M. / Stuart, J.W. / Lam, A.T. / Guenther, R. / Agris, P.F. |  Citation Citation Journal: Nat.Struct.Biol. / Year: 1996 Journal: Nat.Struct.Biol. / Year: 1996Title: Design, biological activity and NMR-solution structure of a DNA analogue of yeast tRNA(Phe) anticodon domain. Authors: Basti, M.M. / Stuart, J.W. / Lam, A.T. / Guenther, R. / Agris, P.F. #1:  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: Direct Observation of Two Base-Pairing Modes of a Cytosine-Thymine Analogue with Guanine in a DNA Z-Form Duplex: Significance for Base Analogue Mutagenesis Authors: Moore, M.H. / Van Meervelt, L. / Salisbury, S.A. / Lin, P.K.T. / Brown, D.M. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  229d.cif.gz 229d.cif.gz | 19.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb229d.ent.gz pdb229d.ent.gz | 13.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  229d.json.gz 229d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  229d_validation.pdf.gz 229d_validation.pdf.gz | 249.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  229d_full_validation.pdf.gz 229d_full_validation.pdf.gz | 250.3 KB | Display | |
| Data in XML |  229d_validation.xml.gz 229d_validation.xml.gz | 2.5 KB | Display | |
| Data in CIF |  229d_validation.cif.gz 229d_validation.cif.gz | 2.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/29/229d https://data.pdbj.org/pub/pdb/validation_reports/29/229d ftp://data.pdbj.org/pub/pdb/validation_reports/29/229d ftp://data.pdbj.org/pub/pdb/validation_reports/29/229d | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 5222.388 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: CHEMICALLY SYNTHESIZED /  Keywords: DEOXYRIBONUCLEIC ACID Keywords: DEOXYRIBONUCLEIC ACID |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other |
|---|
- Processing
Processing
| NMR software | Name: Discover / Developer: BIOSYM TECHNOLOGIES / Classification: refinement |
|---|---|
| Refinement | Software ordinal: 1 Details: NUMBER OF ATOMS USED IN REFINEMENT. NUMBER OF PROTEIN ATOMS 0 NUMBER OF NUCLEIC ACID ATOMS 537 NUMBER OF HETEROGEN ATOMS 0 NUMBER OF SOLVENT ATOMS 0 THE SOLUTION STRUCTURE OF THE 17-MER DNA ...Details: NUMBER OF ATOMS USED IN REFINEMENT. NUMBER OF PROTEIN ATOMS 0 NUMBER OF NUCLEIC ACID ATOMS 537 NUMBER OF HETEROGEN ATOMS 0 NUMBER OF SOLVENT ATOMS 0 THE SOLUTION STRUCTURE OF THE 17-MER DNA HAIRPIN LOOP WAS DETERMINED BASED ON 246 DISTANCE AND DIHEDRAL RESTRAINTS ESTIMATED FROM 2D NOE SPECTRUM. TEN LOW ENERGY STRUCTURES WERE USED TO GENERATE THIS AVERAGE STRUCTURE WHICH WAS FURTHER ENERGY MINIMIZED. |
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller







 PDBj
PDBj







































