+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2bj6 | ||||||
|---|---|---|---|---|---|---|---|
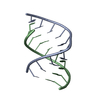
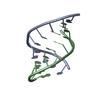
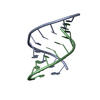
| Title | Crystal Structure of a decameric HNA-RNA hybrid | ||||||
 Components Components |
| ||||||
 Keywords Keywords | NUCLEIC ACID (HNA/RNA) / DNA / RNA / HNA / HEXITOL NUCLEIC ACID / HYBRID / DUPLEX / MODIFIED NUCLEIC ACID / ANTISENSE / BACKBONE MODIFICATION / NUCLEIC ACID (HNA-RNA) complex | ||||||
| Function / homology | DNA / RNA Function and homology information Function and homology information | ||||||
| Biological species |  HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.6 Å MAD / Resolution: 2.6 Å | ||||||
 Authors Authors | Maier, T. / Przylas, I. / Straeter, N. / Herdewijn, P. / Saenger, W. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2005 Journal: J.Am.Chem.Soc. / Year: 2005Title: Reinforced Hna Backbone Hydration in the Crystal Structure of a Decameric Hna/RNA Hybrid Authors: Maier, T. / Przylas, I. / Strater, N. / Herdewijn, P. / Saenger, W. #1:  Journal: J.Am.Chem.Soc. / Year: 1998 Journal: J.Am.Chem.Soc. / Year: 1998Title: Molecular Dynamics Simulation to Inverstigate Differences in Minor Groove Hydration of Hna-RNA Hybrids as Compared to Hna-DNA Complexes Authors: Dewinter, H. / Lescrinier, E. / Vanaerschot, A. / Herdewijn, P. #2:  Journal: Chem.Biol. / Year: 2000 Journal: Chem.Biol. / Year: 2000Title: Solution Structure of a Hna-RNA Hybrid Authors: Lescrinier, E. / Esnouf, R. / Schraml, J. / Busson, R. / Heus, H.A. / Hilbers, C.W. / Herdewijn, P. #3:  Journal: J.Am.Chem.Soc. / Year: 2002 Journal: J.Am.Chem.Soc. / Year: 2002Title: Crystal Structure of Double Helical Hexitol Nucleic Acids Authors: Declercq, R. / Vanaerschot, A. / Read, R.J. / Herdewijn, P. / Vanmeervelt, L. #4:  Journal: Acta Crystallogr.,Sect.C / Year: 1996 Journal: Acta Crystallogr.,Sect.C / Year: 1996Title: 1,5-Anhydro-2,3-Dideoxy-2-(Guanin-9-Yl)-D-Arabino-Hexitol Authors: Declerq, R. / Herdewijn, P. / Vanmeervelt, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2bj6.cif.gz 2bj6.cif.gz | 58.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2bj6.ent.gz pdb2bj6.ent.gz | 48.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2bj6.json.gz 2bj6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bj/2bj6 https://data.pdbj.org/pub/pdb/validation_reports/bj/2bj6 ftp://data.pdbj.org/pub/pdb/validation_reports/bj/2bj6 ftp://data.pdbj.org/pub/pdb/validation_reports/bj/2bj6 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3145.249 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: CHEMICAL SYNTHESIS, PHOSPHOROTHIOATE / Source: (synth.)  HOMO SAPIENS (human) HOMO SAPIENS (human)#2: RNA chain | Mass: 3216.973 Da / Num. of mol.: 4 / Source method: obtained synthetically / Details: CHEMICAL SYNTHESIS, PHOSPHOROTHIOATE / Source: (synth.)  HOMO SAPIENS (human) HOMO SAPIENS (human)#3: Chemical | ChemComp-SO4 / #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.53 Å3/Da / Density % sol: 66.2 % |
|---|---|
| Crystal grow | Details: 0.9M LISO4, 10MM MG(AC)2,0.1M TRIS/HCL, PH 8.0 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: BM30A / Wavelength: 0.92067 / Beamline: BM30A / Wavelength: 0.92067 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Nov 25, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.92067 Å / Relative weight: 1 |
| Reflection | Resolution: 2.54→35 Å / Num. obs: 11676 / % possible obs: 99.4 % / Redundancy: 6.64 % / Rmerge(I) obs: 0.06 / Net I/σ(I): 28.7 |
| Reflection shell | Resolution: 2.54→2.63 Å / Mean I/σ(I) obs: 3.2 / % possible all: 95.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 2.6→35 Å / Data cutoff high absF: 10000 / Cross valid method: THROUGHOUT / σ(F): 0 MAD / Resolution: 2.6→35 Å / Data cutoff high absF: 10000 / Cross valid method: THROUGHOUT / σ(F): 0 Details: DIFFERENT ATOM NUMBERING SCHEMES ARE USED IN THE LITERATURE. PLEASE SEE PRIMARY AND SECONDARY CITATIONS FOR A DETAILED DESCRIPTION. STEREOCHEMICAL PARAMETERS FOR HNA MONOMERS HAVE BEEN ...Details: DIFFERENT ATOM NUMBERING SCHEMES ARE USED IN THE LITERATURE. PLEASE SEE PRIMARY AND SECONDARY CITATIONS FOR A DETAILED DESCRIPTION. STEREOCHEMICAL PARAMETERS FOR HNA MONOMERS HAVE BEEN DERIVED FROM SMALL MOLECULE X-RAY STRUCTURES, SEE SECONDARY REFERENCES
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 48.1 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→35 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.6→2.64 Å / Total num. of bins used: 23 /
|
 Movie
Movie Controller
Controller






 PDBj
PDBj




