+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1xt7 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Daptomycin NMR Structure | ||||||
 Components Components | DAPTOMYCIN | ||||||
 Keywords Keywords | ANTIBIOTIC / DAPTOMYCIN / CUBICIN / LIPOPEPTIDE / CALCIUM-DEPENDENT | ||||||
| Function / homology | Daptomycin / DECANOIC ACID / :  Function and homology information Function and homology information | ||||||
| Biological species |  STREPTOMYCES ROSEOSPORUS (bacteria) STREPTOMYCES ROSEOSPORUS (bacteria) | ||||||
| Method | SOLUTION NMR / CONSTRAINED MOLECULAR DYNAMICS | ||||||
 Authors Authors | Ball, L.-J. / Goult, C.M. / Donarski, J.A. / Micklefield, J. / Ramesh, V. | ||||||
 Citation Citation |  Journal: Org.Biomol.Chem. / Year: 2004 Journal: Org.Biomol.Chem. / Year: 2004Title: NMR Structure Determination and Calcium Binding Effects of Lipopeptide Antibiotic Daptomycin Authors: Ball, L.-J. / Goult, C.M. / Donarski, J.A. / Micklefield, J. / Ramesh, V. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1xt7.cif.gz 1xt7.cif.gz | 13.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1xt7.ent.gz pdb1xt7.ent.gz | 7.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1xt7.json.gz 1xt7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xt/1xt7 https://data.pdbj.org/pub/pdb/validation_reports/xt/1xt7 ftp://data.pdbj.org/pub/pdb/validation_reports/xt/1xt7 ftp://data.pdbj.org/pub/pdb/validation_reports/xt/1xt7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 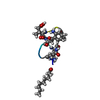
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide |   Type: Cyclic lipopeptide / Class: Antibiotic / Mass: 1484.440 Da / Num. of mol.: 1 / Source method: obtained synthetically Type: Cyclic lipopeptide / Class: Antibiotic / Mass: 1484.440 Da / Num. of mol.: 1 / Source method: obtained syntheticallyDetails: DAPTOMYCIN IS AN ACIDIC CYCLIC LIPOPEPTIDE. THE SCAFFOLD IS MADE OF TWO PARTS: (1) THREE RESIDUES N-TERM EXOCYCLIC PART (2) A DECAPEPTIDE LACTONE RING DERIVED FROM CYCLISATION OF THR3 SIDE ...Details: DAPTOMYCIN IS AN ACIDIC CYCLIC LIPOPEPTIDE. THE SCAFFOLD IS MADE OF TWO PARTS: (1) THREE RESIDUES N-TERM EXOCYCLIC PART (2) A DECAPEPTIDE LACTONE RING DERIVED FROM CYCLISATION OF THR3 SIDE CHAIN ONTO THE C-TER CARBOXYL GROUP THE N-DECANOYL FATTY ACID IS LINKED TO THE MAIN BODY OF THE MOLECULE VIA N-TERM ACYLATION. Source: (synth.)  STREPTOMYCES ROSEOSPORUS (bacteria) / References: NOR: NOR00001, Daptomycin STREPTOMYCES ROSEOSPORUS (bacteria) / References: NOR: NOR00001, Daptomycin |
|---|---|
| #2: Chemical | ChemComp-DKA / 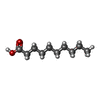  Type: Cyclic lipopeptide / Class: Antibiotic / Mass: 172.265 Da / Num. of mol.: 1 / Source method: obtained synthetically / Formula: C10H20O2 Type: Cyclic lipopeptide / Class: Antibiotic / Mass: 172.265 Da / Num. of mol.: 1 / Source method: obtained synthetically / Formula: C10H20O2Details: DAPTOMYCIN IS AN ACIDIC CYCLIC LIPOPEPTIDE. THE SCAFFOLD IS MADE OF TWO PARTS: (1) THREE RESIDUES N-TERM EXOCYCLIC PART (2) A DECAPEPTIDE LACTONE RING DERIVED FROM CYCLISATION OF THR3 SIDE ...Details: DAPTOMYCIN IS AN ACIDIC CYCLIC LIPOPEPTIDE. THE SCAFFOLD IS MADE OF TWO PARTS: (1) THREE RESIDUES N-TERM EXOCYCLIC PART (2) A DECAPEPTIDE LACTONE RING DERIVED FROM CYCLISATION OF THR3 SIDE CHAIN ONTO THE C-TER CARBOXYL GROUP THE N-DECANOYL FATTY ACID IS LINKED TO THE MAIN BODY OF THE MOLECULE VIA N-TERM ACYLATION. References: Daptomycin |
| Compound details | DAPTOMYCIN IS A CYCLIC TRIDECAMER LIPOPETIDE. HERE, DAPTOMYCIN IS REPRESENTED BY GROUPING TOGETHER ...DAPTOMYCIN |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Type: 2D NOESY |
| NMR details | Text: THIS STRUCTURE WAS DETERMINED USING STANDARD 2D HOMONUCLEAR TECHNIQUES. |
- Sample preparation
Sample preparation
| Details |
| ||||||
|---|---|---|---|---|---|---|---|
| Sample conditions | pH: 5.05 / Pressure: AMBIENT / Temperature: 298 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Bruker DRX / Manufacturer: Bruker / Model: DRX / Field strength: 600 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: CONSTRAINED MOLECULAR DYNAMICS / Software ordinal: 1 Details: A TOTAL OF 52 DISTANCE RESTRAINTS WERE USED FOR NMR STRUCTURE CALCULATION. | ||||||||||||||||||
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller













 PDBj
PDBj