[English] 日本語
 Yorodumi
Yorodumi- PDB-1xj9: Crystal structure of a partly self-complementary peptide nucleic ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1xj9 | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of a partly self-complementary peptide nucleic acid (PNA) oligomer showing a duplex-triplex network | ||||||||||||||||||||||||||
 Components Components | peptide nucleic acid, (H-P(* Keywords KeywordsPEPTIDE NUCLEIC ACID / PNA / partly self-complementary / duplex-triplex complex / right-handed / left-handed | Function / homology | PEP_NUC / PEP_NUC (> 10) |  Function and homology information Function and homology informationBiological species | synthetic construct (others) | Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / crystal 1: single wavelength protocol. crystal 2: MAD protocol with data collected on 5-bromo-uracil derivative crystal at 0.9177, 0.9185, 0.9110, 0.9218 A. the condition for crystal 2 was as follows. collection data: 05-NOV-2000, temperature(kelvin): 110, PH: 4.80, the details of the source of radiation: synchrotron, EMBL/DESY, HAMBURG beamline BW7A / Resolution: 2.6 Å SYNCHROTRON / crystal 1: single wavelength protocol. crystal 2: MAD protocol with data collected on 5-bromo-uracil derivative crystal at 0.9177, 0.9185, 0.9110, 0.9218 A. the condition for crystal 2 was as follows. collection data: 05-NOV-2000, temperature(kelvin): 110, PH: 4.80, the details of the source of radiation: synchrotron, EMBL/DESY, HAMBURG beamline BW7A / Resolution: 2.6 Å  Authors AuthorsPetersson, B. / Nielsen, B.B. / Rasmussen, H. / Larsen, I.K. / Gajhede, M. / Nielsen, P.E. / Kastrup, J.S. |  Citation Citation Journal: J.Am.Chem.Soc. / Year: 2005 Journal: J.Am.Chem.Soc. / Year: 2005Title: Crystal Structure of a Partly Self-Complementary Peptide Nucleic Acid (PNA) Oligomer Showing a Duplex-Triplex Network Authors: Petersson, B. / Nielsen, B.B. / Rasmussen, H. / Larsen, I.K. / Gajhede, M. / Nielsen, P.E. / Kastrup, J.S. #1:  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 A resolution Authors: Rasmussen, H. / Kastrup, J.S. / Nielsen, J.N. / Nielsen, J.M. / Nielsen, P.E. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1xj9.cif.gz 1xj9.cif.gz | 19.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1xj9.ent.gz pdb1xj9.ent.gz | 15.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1xj9.json.gz 1xj9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xj/1xj9 https://data.pdbj.org/pub/pdb/validation_reports/xj/1xj9 ftp://data.pdbj.org/pub/pdb/validation_reports/xj/1xj9 ftp://data.pdbj.org/pub/pdb/validation_reports/xj/1xj9 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
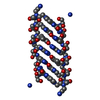
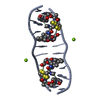
| Details | THIS ENTRY CONTAINS THE CRYSTALLOGRAPHIC ASYMMETRIC UNIT WHICH CONSISTS OF 2 CHAINS. A right-handed duplex (from chain A) is generated with the symmetry operation: -x+2, y, -z+1. / A left-handed duplex (from chain B) is generated with the symmetry operation: -x+1, y, -z |
- Components
Components
| #1: Peptide nucleic acid | Mass: 2866.849 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #2: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
|
-Data collection
| Diffraction | Mean temperature: 293 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: BM14 / Wavelength: 1 Å / Beamline: BM14 / Wavelength: 1 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Feb 17, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.6→30 Å / Num. all: 1839 / Num. obs: 1839 / % possible obs: 98.6 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3 % / Rmerge(I) obs: 0.045 / Net I/σ(I): 22.4 |
| Reflection shell | Resolution: 2.6→2.69 Å / Rmerge(I) obs: 0.183 / Mean I/σ(I) obs: 5.3 / Num. unique all: 172 / % possible all: 95.6 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure: crystal 1: single wavelength protocol. crystal 2: MAD protocol with data collected on 5-bromo-uracil derivative crystal at 0.9177, 0.9185, 0.9110, 0.9218 A. the ...Method to determine structure: crystal 1: single wavelength protocol. crystal 2: MAD protocol with data collected on 5-bromo-uracil derivative crystal at 0.9177, 0.9185, 0.9110, 0.9218 A. the condition for crystal 2 was as follows. collection data: 05-NOV-2000, temperature(kelvin): 110, PH: 4.80, the details of the source of radiation: synchrotron, EMBL/DESY, HAMBURG beamline BW7A Resolution: 2.6→27 Å / Isotropic thermal model: Isotropic / Cross valid method: THROUGHOUT / σ(F): 3 / Stereochemistry target values: Engh & Huber / Details: Own parameter and topology files created
| |||||||||||||||||||||||||
| Displacement parameters | Biso mean: 42.5 Å2 | |||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→27 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.6→2.86 Å / Rfactor Rfree error: 0.092
|
 Movie
Movie Controller
Controller














 PDBj
PDBj
