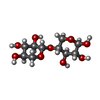
Entry Database : PDB / ID : 1w9tTitle Structure of a beta-1,3-glucan binding CBM6 from Bacillus halodurans in complex with xylobiose BH0236 PROTEIN Keywords / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species BACILLUS HALODURANS (bacteria)Method / / Resolution : 1.62 Å Authors Boraston, A.B. / van Bueren, A.L. Journal : J.Biol.Chem. / Year : 2005Title : Family 6 Carbohydrate Binding Modules Recognize the Non-Reducing End of Beta-1,3-Linked Glucans by Presenting a Unique Ligand Binding SurfaceAuthors : Van Bueren, A.L. / Moreland, C. / Gilbert, H.J. / Boraston, A.B. History Deposition Oct 18, 2004 Deposition site / Processing site Revision 1.0 Nov 3, 2004 Provider / Type Revision 1.1 Aug 7, 2013 Group Derived calculations / Other ... Derived calculations / Other / Refinement description / Structure summary / Version format compliance Revision 2.0 Jul 29, 2020 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Derived calculations / Other / Structure summary Category atom_site / atom_site_anisotrop ... atom_site / atom_site_anisotrop / chem_comp / entity / entity_name_com / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_database_status / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_molecule_features / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / pdbx_struct_conn_angle / pdbx_unobs_or_zero_occ_atoms / pdbx_validate_close_contact / struct_asym / struct_conn / struct_conn_type / struct_site / struct_site_gen Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_asym_id / _atom_site.auth_atom_id / _atom_site.auth_comp_id / _atom_site.auth_seq_id / _atom_site.label_alt_id / _atom_site.label_asym_id / _atom_site.label_atom_id / _atom_site.label_comp_id / _atom_site.label_entity_id / _atom_site.occupancy / _atom_site.type_symbol / _atom_site_anisotrop.U[1][1] / _atom_site_anisotrop.U[1][2] / _atom_site_anisotrop.U[1][3] / _atom_site_anisotrop.U[2][2] / _atom_site_anisotrop.U[2][3] / _atom_site_anisotrop.U[3][3] / _atom_site_anisotrop.pdbx_auth_asym_id / _atom_site_anisotrop.pdbx_auth_atom_id / _atom_site_anisotrop.pdbx_auth_comp_id / _atom_site_anisotrop.pdbx_auth_seq_id / _atom_site_anisotrop.pdbx_label_alt_id / _atom_site_anisotrop.pdbx_label_asym_id / _atom_site_anisotrop.pdbx_label_atom_id / _atom_site_anisotrop.pdbx_label_comp_id / _atom_site_anisotrop.type_symbol / _chem_comp.name / _chem_comp.type / _pdbx_database_status.status_code_sf / _pdbx_entity_nonpoly.comp_id / _pdbx_entity_nonpoly.entity_id / _pdbx_entity_nonpoly.name / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr2_auth_asym_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _pdbx_unobs_or_zero_occ_atoms.auth_atom_id / _pdbx_unobs_or_zero_occ_atoms.label_asym_id / _pdbx_unobs_or_zero_occ_atoms.label_atom_id / _pdbx_validate_close_contact.auth_atom_id_1 / _pdbx_validate_close_contact.auth_atom_id_2 / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_conn_type.id Description / Provider / Type Revision 2.1 Dec 13, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_conn Item _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI ... _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.pdbx_leaving_atom_flag
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information BACILLUS HALODURANS (bacteria)
BACILLUS HALODURANS (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.62 Å
MOLECULAR REPLACEMENT / Resolution: 1.62 Å  Authors
Authors Citation
Citation Journal: J.Biol.Chem. / Year: 2005
Journal: J.Biol.Chem. / Year: 2005 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 1w9t.cif.gz
1w9t.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb1w9t.ent.gz
pdb1w9t.ent.gz PDB format
PDB format 1w9t.json.gz
1w9t.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/w9/1w9t
https://data.pdbj.org/pub/pdb/validation_reports/w9/1w9t ftp://data.pdbj.org/pub/pdb/validation_reports/w9/1w9t
ftp://data.pdbj.org/pub/pdb/validation_reports/w9/1w9t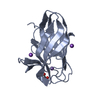


 Links
Links Assembly
Assembly
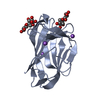
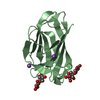
 Components
Components BACILLUS HALODURANS (bacteria) / Production host:
BACILLUS HALODURANS (bacteria) / Production host: 




 X-RAY DIFFRACTION
X-RAY DIFFRACTION Sample preparation
Sample preparation ROTATING ANODE / Wavelength: 1.5418
ROTATING ANODE / Wavelength: 1.5418  Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










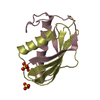

 PDBj
PDBj