+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ul9 | ||||||
|---|---|---|---|---|---|---|---|
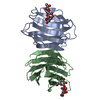
| Title | CGL2 ligandfree | ||||||
 Components Components | galectin-2 | ||||||
 Keywords Keywords | SUGAR BINDING PROTEIN / galectin / lectin / beta-galactoside binding lectin / sugar binding | ||||||
| Function / homology |  Function and homology information Function and homology informationendomembrane system / carbohydrate binding / extracellular region / membrane Similarity search - Function | ||||||
| Biological species |  Coprinopsis cinerea (fungus) Coprinopsis cinerea (fungus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.22 Å MOLECULAR REPLACEMENT / Resolution: 2.22 Å | ||||||
 Authors Authors | Walser, P.J. / Haebel, P.W. / Kuenzler, M. / Kues, U. / Aebi, M. / Ban, N. | ||||||
 Citation Citation |  Journal: STRUCTURE / Year: 2004 Journal: STRUCTURE / Year: 2004Title: Structure and Functional Analysis of the Fungal Galectin CGL2 Authors: Walser, P.J. / Haebel, P.W. / Kuenzler, M. / Sargent, D. / Kues, U. / Aebi, M. / Ban, N. #1:  Journal: ACTA CRYSTALLOGR.,SECT.D / Year: 1998 Journal: ACTA CRYSTALLOGR.,SECT.D / Year: 1998Title: Crystallography & NMR system: A new software suite for macromolecular structure determination. Authors: Brunger, A.T. / Adams, P.D. / Clore, G.M. / DeLano, W.L. / Gros, P. / Grosse-Kunstleve, R.W. / Jiang, J.S. / Kuszewski, J. / Nilges, M. / Pannu, N.S. / Read, R.J. / Rice, L.M. / Simonson, T. / Warren, G.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ul9.cif.gz 1ul9.cif.gz | 76.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ul9.ent.gz pdb1ul9.ent.gz | 58 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ul9.json.gz 1ul9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1ul9_validation.pdf.gz 1ul9_validation.pdf.gz | 429 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1ul9_full_validation.pdf.gz 1ul9_full_validation.pdf.gz | 433.8 KB | Display | |
| Data in XML |  1ul9_validation.xml.gz 1ul9_validation.xml.gz | 16.8 KB | Display | |
| Data in CIF |  1ul9_validation.cif.gz 1ul9_validation.cif.gz | 24.1 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ul/1ul9 https://data.pdbj.org/pub/pdb/validation_reports/ul/1ul9 ftp://data.pdbj.org/pub/pdb/validation_reports/ul/1ul9 ftp://data.pdbj.org/pub/pdb/validation_reports/ul/1ul9 | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | galectin tetramer from crystallographic dimer by -x -y, z |
- Components
Components
| #1: Protein | Mass: 16766.949 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Coprinopsis cinerea (fungus) / Gene: cgl2 / Plasmid: pYADE4 / Production host: Coprinopsis cinerea (fungus) / Gene: cgl2 / Plasmid: pYADE4 / Production host:  #2: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.57 Å3/Da / Density % sol: 51.74 % |
|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 7.3 Details: PEG 3350, PEG 400, sodium phosphate, sodium chloride, pH 7.3, VAPOR DIFFUSION, HANGING DROP, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: May 8, 2003 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.22→43.79 Å / Num. all: 19215 / Num. obs: 19215 / % possible obs: 94.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 5.1 % / Biso Wilson estimate: 18.3 Å2 / Limit h max: 30 / Limit h min: 0 / Limit k max: 51 / Limit k min: 0 / Limit l max: 22 / Limit l min: 0 / Observed criterion F max: 1362778.32 / Observed criterion F min: 2.35 / Rsym value: 0.068 / Net I/σ(I): 21.8 |
| Reflection shell | Resolution: 2.22→2.32 Å / Redundancy: 4.2 % / Mean I/σ(I) obs: 9.8 / Num. unique all: 1334 / Rsym value: 0.141 / % possible all: 66.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: CGL2-lactose Resolution: 2.22→43.79 Å / Rfactor Rfree error: 0.008 / Occupancy max: 1 / Occupancy min: 1 / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: CNS bulk solvent model used / Bsol: 46.2926 Å2 / ksol: 0.371495 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 75.58 Å2 / Biso mean: 21.2 Å2 / Biso min: 7.25 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine Biso |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.22→43.79 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller























 PDBj
PDBj
