[English] 日本語
 Yorodumi
Yorodumi- PDB-1pnv: Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1pnv | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
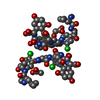
| Title | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and Vancomycin | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | TRANSFERASE/ANTIBIOTIC / GT-B GLYCOSYLTRANSFERASE / ROSSMANN FOLD / GLYCOPEPTIDE / ANTIBIOTIC / TRANSFERASE-ANTIBIOTIC COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroorienticin B synthase / vancomycin biosynthetic process / UDP-glycosyltransferase activity / hexosyltransferase activity / carbohydrate metabolic process Similarity search - Function | |||||||||
| Biological species |  AMYCOLATOPSIS ORIENTALIS (bacteria) AMYCOLATOPSIS ORIENTALIS (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 2.8 Å MAD / Resolution: 2.8 Å | |||||||||
 Authors Authors | Mulichak, A.M. / Losey, H.C. / Lu, W. / Wawrzak, Z. / Walsh, C.T. / Garavito, R.M. | |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2003 Journal: Proc.Natl.Acad.Sci.USA / Year: 2003Title: Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway. Authors: Mulichak, A.M. / Losey, H.C. / Lu, W. / Wawrzak, Z. / Walsh, C.T. / Garavito, R.M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1pnv.cif.gz 1pnv.cif.gz | 157.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1pnv.ent.gz pdb1pnv.ent.gz | 122.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1pnv.json.gz 1pnv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pn/1pnv https://data.pdbj.org/pub/pdb/validation_reports/pn/1pnv ftp://data.pdbj.org/pub/pdb/validation_reports/pn/1pnv ftp://data.pdbj.org/pub/pdb/validation_reports/pn/1pnv | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological assembly is a monomer. |
- Components
Components
| #1: Protein | Mass: 42773.398 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  AMYCOLATOPSIS ORIENTALIS (bacteria) / Gene: GTFA / Plasmid: PET22B / Production host: AMYCOLATOPSIS ORIENTALIS (bacteria) / Gene: GTFA / Plasmid: PET22B / Production host:  #2: Protein/peptide | | 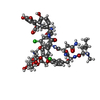  Type: Glycopeptide / Class: Antibiotic / Mass: 1149.977 Da / Num. of mol.: 1 / Source method: isolated from a natural source Type: Glycopeptide / Class: Antibiotic / Mass: 1149.977 Da / Num. of mol.: 1 / Source method: isolated from a natural sourceDetails: VANCOMYCIN IS A TRICYCLIC GLYCOPEPTIDE, GLYCOSYLATED BY A DISACCHARIDE (RESIDUES 8 AND 9) ON RESIDUE 4. Source: (natural)  AMYCOLATOPSIS ORIENTALIS (bacteria) / References: NOR: NOR00681, Vancomycin AMYCOLATOPSIS ORIENTALIS (bacteria) / References: NOR: NOR00681, Vancomycin#3: Polysaccharide | vancosamine-(1-2)-beta-D-glucopyranose | #4: Chemical | ChemComp-TYD / | #5: Water | ChemComp-HOH / | Compound details | VANCOMYCIN IS A TRICYCLIC GLYCOPEPTIDE. THE SCAFFOLD IS A HEPTAPEPTIDE WITH THE CONFIGURATION D-D-L- ...VANCOMYCIN | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.98 Å3/Da / Density % sol: 69.1 % |
|---|---|
| Crystal grow | pH: 6.1 Details: NA,K PHOSPHATE, PH 6.1, VAPOR DIFFUSION, HANGING DROP, TEMPERATURE 298K |
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop |
| Components of the solutions | *PLUS Conc.: 1.3 M / Common name: sodium potassium phosphate / Details: pH6.1 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 5ID-B / Wavelength: 1 / Beamline: 5ID-B / Wavelength: 1 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Nov 2, 2002 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.7→30 Å / Num. obs: 36772 / % possible obs: 97.4 % / Observed criterion σ(I): -3 / Redundancy: 5.5 % / Rsym value: 0.072 / Net I/σ(I): 12.6 |
| Reflection shell | Resolution: 2.7→2.8 Å / Rsym value: 0.236 / % possible all: 98.3 |
| Reflection | *PLUS Highest resolution: 2.8 Å / Lowest resolution: 30 Å / Num. obs: 30058 / % possible obs: 98.3 % / Rmerge(I) obs: 0.072 |
| Reflection shell | *PLUS Rmerge(I) obs: 0.236 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MAD / Resolution: 2.8→30 Å / Cross valid method: THROUGHOUT / σ(F): 2 / Stereochemistry target values: ENGH & HUBER MAD / Resolution: 2.8→30 Å / Cross valid method: THROUGHOUT / σ(F): 2 / Stereochemistry target values: ENGH & HUBER
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 62.8 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.36 Å / Luzzati d res low obs: 5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.8→30 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS % reflection Rfree: 5 % / Rfactor Rfree: 0.274 / Rfactor Rwork: 0.228 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller





















 PDBj
PDBj