[English] 日本語
 Yorodumi
Yorodumi- PDB-1ne7: HUMAN GLUCOSAMINE-6-PHOSPHATE DEAMINASE ISOMERASE AT 1.75 A RESOL... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ne7 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HUMAN GLUCOSAMINE-6-PHOSPHATE DEAMINASE ISOMERASE AT 1.75 A RESOLUTION COMPLEXED WITH N-ACETYL-GLUCOSAMINE-6-PHOSPHATE AND 2-DEOXY-2-AMINO-GLUCITOL-6-PHOSPHATE | ||||||||||||
 Components Components | Glucosamine-6-phosphate isomerase | ||||||||||||
 Keywords Keywords | HYDROLASE / V-TYPE LIKE ALLOSTERIC ENZYME / CONFORMATIONAL DISORDER / CONFORMATIONAL DIFFERENCES | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationglucosamine catabolic process / glucosamine-6-phosphate deaminase / glucosamine-6-phosphate deaminase activity / N-acetylglucosamine catabolic process / N-acetylneuraminate catabolic process / UDP-N-acetylglucosamine biosynthetic process / Glycolysis / single fertilization / isomerase activity / generation of precursor metabolites and energy ...glucosamine catabolic process / glucosamine-6-phosphate deaminase / glucosamine-6-phosphate deaminase activity / N-acetylglucosamine catabolic process / N-acetylneuraminate catabolic process / UDP-N-acetylglucosamine biosynthetic process / Glycolysis / single fertilization / isomerase activity / generation of precursor metabolites and energy / carbohydrate metabolic process / extracellular exosome / identical protein binding / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.75 Å MOLECULAR REPLACEMENT / Resolution: 1.75 Å | ||||||||||||
 Authors Authors | Arreola, R. / Valderrama, B. / Morante, M.L. / Horjales, E. | ||||||||||||
 Citation Citation |  Journal: Febs Lett. / Year: 2003 Journal: Febs Lett. / Year: 2003Title: Two mammalian glucosamine-6-phosphate deaminases: a structural and genetic study. Authors: Arreola, R. / Valderrama, B. / Morante, M.L. / Horjales, E. #1:  Journal: BIOCHIM.BIOPHYS.ACTA / Year: 1998 Journal: BIOCHIM.BIOPHYS.ACTA / Year: 1998Title: Glucosamine-6-phosphate deaminase from beef kidney is an allosteric system of the V-type. Authors: Lara-Lemus, R. / Calcagno, M.L. #2:  Journal: Structure / Year: 1995 Journal: Structure / Year: 1995Title: Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution. Authors: Oliva, G. / Fontes, M.R. / Garratt, R.C. / Altamirano, M.M. / Calcagno, M.L. / Horjales, E. | ||||||||||||
| History |
| ||||||||||||
| Remark 600 | HETEROGEN THE LIGANDS SO4 AND AGP ARE IN ALTERNATE POSITIONS. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ne7.cif.gz 1ne7.cif.gz | 396.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ne7.ent.gz pdb1ne7.ent.gz | 319.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ne7.json.gz 1ne7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ne/1ne7 https://data.pdbj.org/pub/pdb/validation_reports/ne/1ne7 ftp://data.pdbj.org/pub/pdb/validation_reports/ne/1ne7 ftp://data.pdbj.org/pub/pdb/validation_reports/ne/1ne7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1deaS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological assembly is the hexamer in the asymmetric unit reported in this entry. Chains: A, B, C, D, E & F. |
- Components
Components
-Protein , 1 types, 6 molecules ABCDEF
| #1: Protein | Mass: 32712.506 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNPI OR HLN OR KIAA0060 / Production host: Homo sapiens (human) / Gene: GNPI OR HLN OR KIAA0060 / Production host:  References: UniProt: P46926, glucosamine-6-phosphate deaminase |
|---|
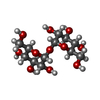
-Sugars , 2 types, 12 molecules 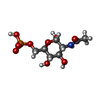
| #2: Polysaccharide | alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose #3: Sugar | ChemComp-16G / |
|---|
-Non-polymers , 3 types, 2207 molecules 
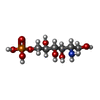



| #4: Chemical | ChemComp-SO4 / #5: Chemical | ChemComp-AGP / #6: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.81 Å3/Da / Density % sol: 56.17 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 291 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 1.68 M ammonium sulphate, 10 mM N-acetyl-glucosamine-6-phosphate and 0.1 mM 2-deoxy-2-amino D-glucitol 6-phosphate, pH 7.0, VAPOR DIFFUSION, SITTING DROP, temperature 291K | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / pH: 7 / Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 93 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-1 / Wavelength: 0.78 Å / Beamline: BL9-1 / Wavelength: 0.78 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Mar 18, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.78 Å / Relative weight: 1 |
| Reflection | Resolution: 1.75→19.97 Å / Num. all: 221759 / Num. obs: 209143 / % possible obs: 94.3 % / Observed criterion σ(F): 1 / Observed criterion σ(I): 0 / Redundancy: 3.8 % / Biso Wilson estimate: 19.5 Å2 / Rsym value: 0.052 / Net I/σ(I): 9.9 |
| Reflection shell | Resolution: 1.75→1.86 Å / Redundancy: 3.6 % / Mean I/σ(I) obs: 3.4 / Num. unique all: 32182 / Rsym value: 0.224 / % possible all: 95.8 |
| Reflection | *PLUS Rmerge(I) obs: 0.052 |
| Reflection shell | *PLUS % possible obs: 96.8 % / Rmerge(I) obs: 0.224 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1DEA Resolution: 1.75→19.97 Å / Rfactor Rfree error: 0.002 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 67.2697 Å2 / ksol: 0.376291 e/Å3 | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 25.9 Å2
| ||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.75→19.97 Å
| ||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.75→1.76 Å / Rfactor Rfree error: 0.004 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS % reflection Rfree: 10 % | ||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Lowest resolution: 1.86 Å / Rfactor Rfree: 0.245 / Rfactor Rwork: 0.223 |
 Movie
Movie Controller
Controller















 PDBj
PDBj