[English] 日本語
 Yorodumi
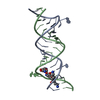
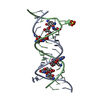
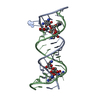
Yorodumi- PDB-1mwl: Crystal structure of geneticin bound to the eubacterial 16S rRNA ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1mwl | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of geneticin bound to the eubacterial 16S rRNA A site | ||||||||||||||||||
 Components Components | 5'-R(* Keywords KeywordsRNA / AMINOGLYCOSIDE ANTIBIOTIC / GENETICIN-A-SITE COMPLEX / 16S RIBOSOMAL RNA / Bulged adenines / UoU pairs | Function / homology | GENETICIN / RNA / RNA (> 10) |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.4 Å MOLECULAR REPLACEMENT / Resolution: 2.4 Å  Authors AuthorsVicens, Q. / Westhof, E. |  Citation Citation Journal: J.Mol.Biol. / Year: 2003 Journal: J.Mol.Biol. / Year: 2003Title: Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide Authors: Vicens, Q. / Westhof, E. #1:  Journal: Chem.Biol. / Year: 2002 Journal: Chem.Biol. / Year: 2002Title: Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding A site Authors: Vicens, Q. / Westhof, E. #2:  Journal: Structure / Year: 2001 Journal: Structure / Year: 2001Title: Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site Authors: Vicens, Q. / Westhof, E. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1mwl.cif.gz 1mwl.cif.gz | 37 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1mwl.ent.gz pdb1mwl.ent.gz | 26.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1mwl.json.gz 1mwl.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1mwl_validation.pdf.gz 1mwl_validation.pdf.gz | 1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1mwl_full_validation.pdf.gz 1mwl_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  1mwl_validation.xml.gz 1mwl_validation.xml.gz | 4.8 KB | Display | |
| Data in CIF |  1mwl_validation.cif.gz 1mwl_validation.cif.gz | 6 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mw/1mwl https://data.pdbj.org/pub/pdb/validation_reports/mw/1mwl ftp://data.pdbj.org/pub/pdb/validation_reports/mw/1mwl ftp://data.pdbj.org/pub/pdb/validation_reports/mw/1mwl | HTTPS FTP |
-Related structure data
| Related structure data |  1lc4S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: RNA chain | Mass: 7048.259 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: eubacterial 16S rRNA A site. The oligonucleotide contains two A sites. #2: Chemical | #3: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.73 Å3/Da / Density % sol: 54.91 % | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 310 K / Method: vapor diffusion, hanging drop / pH: 6.4 Details: MPD, MAGNESIUM SULPHATE, POTASSIUM CHLORIDE, SODIUM CHLORIDE, SODIUM CACODYLATE, pH 6.4, VAPOR DIFFUSION, HANGING DROP, temperature 310K | |||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 37 ℃ | |||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| ||||||||||||||||||
| Detector |
| ||||||||||||||||||
| Radiation |
| ||||||||||||||||||
| Radiation wavelength | Wavelength: 0.934 Å / Relative weight: 1 | ||||||||||||||||||
| Reflection | Resolution: 2.4→25 Å / Num. obs: 5466 / % possible obs: 88.8 % / Redundancy: 4.8 % / Biso Wilson estimate: 65.5 Å2 / Rsym value: 0.062 / Net I/σ(I): 39.3 | ||||||||||||||||||
| Reflection shell | Resolution: 2.4→2.49 Å / Mean I/σ(I) obs: 6.7 / Num. unique all: 566 / Rsym value: 0.211 / % possible all: 92.8 | ||||||||||||||||||
| Reflection | *PLUS Num. measured all: 24145 / Rmerge(I) obs: 0.062 | ||||||||||||||||||
| Reflection shell | *PLUS Highest resolution: 2.4 Å / % possible obs: 92.8 % / Num. unique obs: 566 / Rmerge(I) obs: 0.211 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1LC4 (RNA ONLY) Resolution: 2.4→22.61 Å / Rfactor Rfree error: 0.014 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 1.5 Stereochemistry target values: G. Parkinson, J. Vojtechovsky, L. Clowney, A.T. Brunger, H.M. Berman, New parameters for the refinement of nucleic acid containing structures, Acta Cryst, D, 52, 57-64 (1996)
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 36.9452 Å2 / ksol: 0.314673 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 57.4 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error free: 0.39 Å / Luzzati sigma a free: 0.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→22.61 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.4→2.49 Å / Rfactor Rfree error: 0.119 / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 25 Å / Rfactor Rfree: 0.25 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Highest resolution: 2.4 Å |
 Movie
Movie Controller
Controller




 PDBj
PDBj

































