[English] 日本語
 Yorodumi
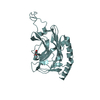
Yorodumi- PDB-1lvm: CATALYTICALLY ACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1lvm | ||||||
|---|---|---|---|---|---|---|---|
| Title | CATALYTICALLY ACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH PRODUCT | ||||||
 Components Components |
| ||||||
 Keywords Keywords | VIRAL PROTEIN / Beta Barrel / Chymotrypsin-type Cystein Protease / Enzyme-peptide Complex | ||||||
| Function / homology |  Function and homology information Function and homology informationnuclear-inclusion-a endopeptidase / helper-component proteinase / hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides / host cell cytoplasmic vesicle / helical viral capsid / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / serine-type peptidase activity / helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / symbiont-mediated suppression of host innate immune response ...nuclear-inclusion-a endopeptidase / helper-component proteinase / hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides / host cell cytoplasmic vesicle / helical viral capsid / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / serine-type peptidase activity / helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / symbiont-mediated suppression of host innate immune response / viral translational frameshifting / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / host cell nucleus / structural molecule activity / proteolysis / RNA binding / ATP binding Similarity search - Function | ||||||
| Biological species |  Tobacco etch virus Tobacco etch virus | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Phan, J. / Zdanov, A. / Evdokimov, A.G. / Tropea, J.E. / Peters III, H.K. / Kapust, R.B. / Li, M. / Wlodawer, A. / Waugh, D.S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2002 Journal: J.Biol.Chem. / Year: 2002Title: Structural basis for the substrate specificity of tobacco etch virus protease. Authors: Phan, J. / Zdanov, A. / Evdokimov, A.G. / Tropea, J.E. / Peters III, H.K. / Kapust, R.B. / Li, M. / Wlodawer, A. / Waugh, D.S. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE AUTHOR STATES RESIDUES 308-310 ARE NOT IN THE CRYSTAL OF CHAINS C AND D AND RESIDUES 309- ...SEQUENCE AUTHOR STATES RESIDUES 308-310 ARE NOT IN THE CRYSTAL OF CHAINS C AND D AND RESIDUES 309-310 ARE NOT IN THE PEPTIDE SEQUENCE FOR THIS MUTANT. RESIDUE GLY 308 WAS CLEAVED OFF BY THE ENZYME AND NOT PRESENT IN THE CRYSTAL. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1lvm.cif.gz 1lvm.cif.gz | 119.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1lvm.ent.gz pdb1lvm.ent.gz | 92.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1lvm.json.gz 1lvm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lv/1lvm https://data.pdbj.org/pub/pdb/validation_reports/lv/1lvm ftp://data.pdbj.org/pub/pdb/validation_reports/lv/1lvm ftp://data.pdbj.org/pub/pdb/validation_reports/lv/1lvm | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1lvbSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 26147.586 Da / Num. of mol.: 2 / Fragment: Residues 1-221 / Mutation: S219D Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Tobacco etch virus / Genus: Potyvirus / Plasmid: BL21(DE3) / Production host: Tobacco etch virus / Genus: Potyvirus / Plasmid: BL21(DE3) / Production host:  #2: Protein/peptide | Mass: 1084.137 Da / Num. of mol.: 2 / Fragment: Residues 302-310 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Tobacco etch virus / Genus: Potyvirus / Plasmid: BL21(DE3) / Production host: Tobacco etch virus / Genus: Potyvirus / Plasmid: BL21(DE3) / Production host:  #3: Protein/peptide | | Mass: 805.896 Da / Num. of mol.: 1 / Fragment: Residues 230-236 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Tobacco etch virus / Genus: Potyvirus / Plasmid: BL21(DE3) / Production host: Tobacco etch virus / Genus: Potyvirus / Plasmid: BL21(DE3) / Production host:  #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.36 Å3/Da / Density % sol: 47.9 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: ammonium sulfate, magnesium chloride, Tris-HCl, pH 8.5, VAPOR DIFFUSION, HANGING DROP, temperature 298K | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X9B / Wavelength: 0.92 Å / Beamline: X9B / Wavelength: 0.92 Å |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Jan 18, 2001 / Details: Mirrors |
| Radiation | Monochromator: Double Crystal / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.92 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→25 Å / Num. all: 49988 / Num. obs: 48452 / % possible obs: 97 % / Observed criterion σ(F): 2 / Observed criterion σ(I): 2 / Redundancy: 6.2 % / Biso Wilson estimate: 15 Å2 / Rmerge(I) obs: 0.058 |
| Reflection shell | Resolution: 1.8→1.86 Å / % possible all: 99.3 |
| Reflection | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 30 Å / Num. obs: 49988 / % possible obs: 99.8 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1LVB Resolution: 1.8→25 Å / Rfactor Rfree error: 0.003 / Data cutoff high absF: 140622.42 / Data cutoff high rms absF: 140622.42 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 54.5232 Å2 / ksol: 0.334191 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.8 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→25 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.91 Å / Rfactor Rfree error: 0.011 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 30 Å / Num. reflection obs: 43567 / Rfactor Rfree: 0.229 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller









 PDBj
PDBj
