[English] 日本語
 Yorodumi
Yorodumi- PDB-1lod: INTERACTION OF A LEGUME LECTIN WITH TWO COMPONENTS OF THE BACTERI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1lod | ||||||
|---|---|---|---|---|---|---|---|
| Title | INTERACTION OF A LEGUME LECTIN WITH TWO COMPONENTS OF THE BACTERIAL CELL WALL | ||||||
 Components Components | (LEGUME ISOLECTIN I ...) x 2 | ||||||
 Keywords Keywords | LECTIN | ||||||
| Function / homology |  Function and homology information Function and homology information | ||||||
| Biological species |  Lathyrus ochrus (yellow-flowered pea) Lathyrus ochrus (yellow-flowered pea) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2.05 Å X-RAY DIFFRACTION / Resolution: 2.05 Å | ||||||
 Authors Authors | Bourne, Y. / Cambillau, C. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 1994 Journal: J.Biol.Chem. / Year: 1994Title: Interaction of a legume lectin with two components of the bacterial cell wall. A crystallographic study. Authors: Bourne, Y. / Ayouba, A. / Rouge, P. / Cambillau, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1lod.cif.gz 1lod.cif.gz | 193.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1lod.ent.gz pdb1lod.ent.gz | 153.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1lod.json.gz 1lod.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lo/1lod https://data.pdbj.org/pub/pdb/validation_reports/lo/1lod ftp://data.pdbj.org/pub/pdb/validation_reports/lo/1lod ftp://data.pdbj.org/pub/pdb/validation_reports/lo/1lod | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 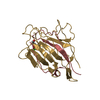
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 6 | 
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: ALA A 80 - ASP A 81 OMEGA = 349.60 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 2: ALA C 80 - ASP C 81 OMEGA = 347.69 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 3: ALA E 80 - ASP E 81 OMEGA = 346.68 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 4: ALA G 80 - ASP G 81 OMEGA = 343.81 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION | ||||||||
| Details | THIS CRYSTAL FORM CONTAINS TWO DIMERS IN THE ASYMMETRIC UNIT. EACH MONOMER CONSISTS OF TWO SEPARATE POLYPEPTIDE CHAINS, ALPHA AND BETA. THE ALPHA CHAIN CONSISTS OF 181 RESIDUES AND THE BETA CHAIN CONSISTS OF 52 RESIDUES. THE BETA AND ALPHA CHAINS OF MONOMER 1 HAVE BEEN ASSIGNED *A* AND *B*. THE BETA AND ALPHA CHAINS OF MONOMER 2 HAVE BEEN ASSIGNED *C* AND *D*. THE SAME ASSIGNMENT HAS BEEN MADE FOR THE SECOND MOLECULE WITH CHAIN IDENTIFIERS *E*, *F*, *G*, *H*. EACH DIMER HAS THE TWO MONOMERS WHICH ARE RELATED BY A NON-CRYSTALLOGRAPHIC TWO-FOLD AXIS. |
- Components
Components
-LEGUME ISOLECTIN I ... , 2 types, 8 molecules ACEGBDFH
| #1: Protein | Mass: 19847.857 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Lathyrus ochrus (yellow-flowered pea) / Organ: SEED / References: UniProt: P04122 Lathyrus ochrus (yellow-flowered pea) / Organ: SEED / References: UniProt: P04122#2: Protein | Mass: 5783.322 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Lathyrus ochrus (yellow-flowered pea) / Organ: SEED / References: UniProt: P12306 Lathyrus ochrus (yellow-flowered pea) / Organ: SEED / References: UniProt: P12306 |
|---|
-Sugars , 1 types, 4 molecules 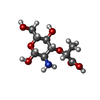
| #5: Sugar | ChemComp-MUR / |
|---|
-Non-polymers , 3 types, 320 molecules 




| #3: Chemical | ChemComp-CA / #4: Chemical | ChemComp-MN / #6: Water | ChemComp-HOH / | |
|---|
-Details
| Sequence details | SEQUENCE ADVISORY NOTICE: DIFFERENCE BETWEEN SWISS-PROT AND PDB SEQUENCE. SWISS-PROT ENTRY NAME: ...SEQUENCE ADVISORY NOTICE: DIFFERENCE |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.41 Å3/Da / Density % sol: 48.98 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Temperature: 20 ℃ / pH: 7.5 / Method: vapor diffusion, sitting dropDetails: Bourne, Y., (1990) Proteins Struct. Funct. Genet., 8, 365. | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | *PLUS Highest resolution: 2.05 Å / Lowest resolution: 6 Å / Num. obs: 56334 / % possible obs: 95 % / Num. measured all: 224984 / Rmerge(I) obs: 0.073 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.05→6 Å / Rfactor Rwork: 0.197 / Rfactor obs: 0.197 / σ(F): 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.05→6 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Num. reflection obs: 55620 / Rfactor obs: 0.197 / Rfactor Rwork: 0.197 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller















 PDBj
PDBj




