[English] 日本語
 Yorodumi
Yorodumi- PDB-1jq9: Crystal structure of a complex formed between phospholipase A2 fr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1jq9 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Phe-Leu-Ser-Tyr-Lys at 1.8 resolution | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/HYDROLASE INHIBITOR / phospholipase A2 / Daboia russelli pulchella / neurotoxic / designed peptide / HYDROLASE / HYDROLASE-HYDROLASE INHIBITOR COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationphospholipase A2 / : / arachidonate secretion / lipid catabolic process / negative regulation of T cell proliferation / phospholipid metabolic process / phospholipid binding / toxin activity / calcium ion binding / extracellular region Similarity search - Function | ||||||
| Biological species |  Daboia russellii pulchella (snake) Daboia russellii pulchella (snake) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.8 Å MOLECULAR REPLACEMENT / Resolution: 1.8 Å | ||||||
 Authors Authors | Chandra, V. / Jasti, J. / Kaur, P. / Dey, S. / Betzel, C. / Singh, T.P. | ||||||
 Citation Citation |  Journal: J.BIOL.CHEM. / Year: 2002 Journal: J.BIOL.CHEM. / Year: 2002Title: Crystal Structure of a Complex Formed between a Snake Venom Phospholipase A2 and a Potent Peptide Inhibitor Phe-Leu-Ser-Tyr-Lys at 1.8 A Resolution Authors: Chandra, V. / Jasti, J. / Kaur, P. / Dey, S. / Perbandt, M. / Srinivasan, A. / Betzel, C. / Singh, T.P. #1:  Journal: J.Mol.Biol. / Year: 2000 Journal: J.Mol.Biol. / Year: 2000Title: Three-dimensional structure of a presynaptic neurotoxic phospholipase A2 from Daboia russelli pulchella at 2.4 resolution Authors: Chandra, V. / Kaur, P. / SRINIVASAN, A. / Singh, T.P. #2:  Journal: Acta Crystallogr.,Sect.D / Year: 2001 Journal: Acta Crystallogr.,Sect.D / Year: 2001Title: Regulation of catalytic function by molecular association: structure of phospholipase A2 from Daboia russelli pulchella (DPLA2) at 1.9 A resolution Authors: Chandra, V. / Kaur, P. / Jasti, J. / Betzel, C. / Singh, T.P. #3:  Journal: J.MOL.BIOL. / Year: 2002 Journal: J.MOL.BIOL. / Year: 2002Title: First structural evidence of a specific inhibition of phospholipase A2 by alpha-tocopherol (vitamin E) and its implications in inflammation: crystal structure of the complex formed between ...Title: First structural evidence of a specific inhibition of phospholipase A2 by alpha-tocopherol (vitamin E) and its implications in inflammation: crystal structure of the complex formed between phospholipase A2 and alpha-tocopherol at 1.8 A resolution Authors: Chandra, V. / Jasti, J. / Kaur, P. / Betzel, C. / Srinivasan, A. / Singh, T.P. #4:  Journal: Biochemistry / Year: 2002 Journal: Biochemistry / Year: 2002Title: Structural basis of phospholipase A2 inhibition for the synthesis of prostaglandins by the plant alkaloid aristolochic acid from a 1.7 A crystal structure Authors: Chandra, V. / Jasti, J. / Kaur, P. / Srinivasan, A. / Betzel, C. / Singh, T.P. #5:  Journal: Acta Crystallogr.,Sect.D / Year: 2002 Journal: Acta Crystallogr.,Sect.D / Year: 2002Title: Design of specific peptide inhibitors of phospholipase A2: structure of a complex formed between Russell's viper phospholipase A2 and a designed peptide Leu-Ala-Ile-Tyr-Ser (LAIYS) Authors: Chandra, V. / Jasti, J. / Kaur, P. / Dey, S. / Srinivasan, A. / Betzel, C. / Singh, T.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1jq9.cif.gz 1jq9.cif.gz | 69.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1jq9.ent.gz pdb1jq9.ent.gz | 51 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1jq9.json.gz 1jq9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jq/1jq9 https://data.pdbj.org/pub/pdb/validation_reports/jq/1jq9 ftp://data.pdbj.org/pub/pdb/validation_reports/jq/1jq9 ftp://data.pdbj.org/pub/pdb/validation_reports/jq/1jq9 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1fe7 S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 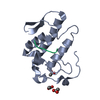
| ||||||||
| 2 | 
| ||||||||
| 3 |
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Details | The two protein chains (A and B) represent the two molecules in the asymmetric unit, with the A molecule containing peptide inhibitor |
- Components
Components
| #1: Protein | Mass: 13629.767 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Daboia russellii pulchella (snake) / Species: Daboia russellii / Strain: pulchella / References: UniProt: P59071, phospholipase A2 Daboia russellii pulchella (snake) / Species: Daboia russellii / Strain: pulchella / References: UniProt: P59071, phospholipase A2#2: Protein/peptide | | Mass: 657.778 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: THE SEQUENCE WAS CHEMICALLY SYNTHESIZED. #3: Chemical | ChemComp-ACY / #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.53 Å3/Da / Density % sol: 51.4 % | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 20mM Sodium cacodylate, 1.4M Ammonium sulfate, 4mM Calcium chloride, 3% dioxane, pH 6.5, VAPOR DIFFUSION, HANGING DROP, temperature 298K | ||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 25 ℃ | ||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 180 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X11 / Wavelength: 0.98 Å / Beamline: X11 / Wavelength: 0.98 Å |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jun 20, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 1.8→20 Å / Num. all: 239988 / Num. obs: 239988 / % possible obs: 94.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.4 % / Biso Wilson estimate: 23.9 Å2 / Rmerge(I) obs: 0.082 / Rsym value: 0.04 / Net I/σ(I): 9.44 |
| Reflection shell | Resolution: 1.8→1.84 Å / Redundancy: 2.4 % / Rmerge(I) obs: 4.7 / Mean I/σ(I) obs: 9.9 / Num. unique all: 1581 / Rsym value: 0.09 / % possible all: 98 |
| Reflection | *PLUS Num. obs: 22959 / Num. measured all: 239988 / Rmerge(I) obs: 0.04 |
| Reflection shell | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 1.86 Å / % possible obs: 98 % / Rmerge(I) obs: 0.09 / Mean I/σ(I) obs: 3.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1FE7  1fe7 Resolution: 1.8→11.95 Å / Rfactor Rfree error: 0.007 / Data cutoff high absF: 2042260.78 / Data cutoff high rms absF: 0 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber / Details: Used weighted full matrix least squares procedure.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 66.0477 Å2 / ksol: 0.332842 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 32.7 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→11.95 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.8→1.84 Å / Rfactor Rfree error: 0.015 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 1.8 Å / Lowest resolution: 20 Å / % reflection Rfree: 5 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj






