[English] 日本語
 Yorodumi
Yorodumi- PDB-1izi: Inhibitor of HIV protease with unusual binding mode potently inhi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1izi | ||||||
|---|---|---|---|---|---|---|---|
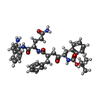
| Title | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | ||||||
 Components Components | proteinase | ||||||
 Keywords Keywords | HYDROLASE / HIV-1 proteinase / triple mutant / potent inhibitor / subsite binding | ||||||
| Function / homology |  Function and homology information Function and homology informationHIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / RNA-directed DNA polymerase / establishment of integrated proviral latency / RNA stem-loop binding ...HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / RNA-directed DNA polymerase / establishment of integrated proviral latency / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / host cell / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding / membrane Similarity search - Function | ||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.15 Å MOLECULAR REPLACEMENT / Resolution: 2.15 Å | ||||||
 Authors Authors | Weber, J. / Mesters, J.R. / Lepsik, M. / Prejdova, J. / Svec, M. / Sponarova, J. / Mlcochova, P. / Skalicka, K. / Strisovsky, K. / Uhlikova, T. ...Weber, J. / Mesters, J.R. / Lepsik, M. / Prejdova, J. / Svec, M. / Sponarova, J. / Mlcochova, P. / Skalicka, K. / Strisovsky, K. / Uhlikova, T. / Soucek, M. / Machala, L. / Stankova, M. / Vondrasek, J. / Klimkait, T. / Kraeusslich, H.-G. / Hilgenfeld, R. / Konvalinka, J. | ||||||
 Citation Citation |  Journal: J.MOL.BIOL. / Year: 2002 Journal: J.MOL.BIOL. / Year: 2002Title: Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains Authors: Weber, J. / Mesters, J.R. / Lepsik, M. / Prejdova, J. / Svec, M. / Sponarova, J. / Mlcochova, P. / Skalicka, K. / Strisovsky, K. / Uhlikova, T. / Soucek, M. / Machala, L. / Stankova, M. / ...Authors: Weber, J. / Mesters, J.R. / Lepsik, M. / Prejdova, J. / Svec, M. / Sponarova, J. / Mlcochova, P. / Skalicka, K. / Strisovsky, K. / Uhlikova, T. / Soucek, M. / Machala, L. / Stankova, M. / Vondrasek, J. / Klimkait, T. / Kraeusslich, H.-G. / Hilgenfeld, R. / Konvalinka, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
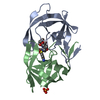
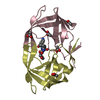
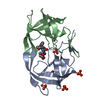
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1izi.cif.gz 1izi.cif.gz | 53.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1izi.ent.gz pdb1izi.ent.gz | 37.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1izi.json.gz 1izi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/iz/1izi https://data.pdbj.org/pub/pdb/validation_reports/iz/1izi ftp://data.pdbj.org/pub/pdb/validation_reports/iz/1izi ftp://data.pdbj.org/pub/pdb/validation_reports/iz/1izi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1izhSC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological assembly is the homodimer as found in the asymmetric unit |
- Components
Components
| #1: Protein | Mass: 10846.781 Da / Num. of mol.: 2 / Mutation: A71V, V82T, I84V Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Human immunodeficiency virus 1 / Genus: Lentivirus / Plasmid: pET24a / Species (production host): Escherichia coli / Production host: Human immunodeficiency virus 1 / Genus: Lentivirus / Plasmid: pET24a / Species (production host): Escherichia coli / Production host:  References: UniProt: Q90EB9, UniProt: P03367*PLUS, HIV-1 retropepsin #2: Chemical | #3: Chemical | ChemComp-Q50 / {( | #4: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.1 Å3/Da / Density % sol: 41.5 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 294 K / Method: vapor diffusion, hanging drop / pH: 5 Details: MES, EDTA, DTT, NaCl, pH 5, VAPOR DIFFUSION, HANGING DROP, temperature 294K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / pH: 6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: X13 / Wavelength: 0.802 Å / Beamline: X13 / Wavelength: 0.802 Å |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Aug 10, 2001 |
| Radiation | Monochromator: SAGITALLY FOCUSED Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.802 Å / Relative weight: 1 |
| Reflection | Resolution: 2.15→40 Å / Num. all: 9695 / Num. obs: 9695 / % possible obs: 98.9 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 5.62 % / Biso Wilson estimate: 33.5 Å2 / Rmerge(I) obs: 0.06 / Net I/σ(I): 19.76 |
| Reflection shell | Resolution: 2.15→2.23 Å / Rmerge(I) obs: 0.323 / Num. unique all: 952 / % possible all: 99.9 |
| Reflection | *PLUS Highest resolution: 2.1 Å / Rmerge(I) obs: 0.06 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1IZH Resolution: 2.15→53.45 Å / Cor.coef. Fo:Fc: 0.959 / Cor.coef. Fo:Fc free: 0.923 / SU B: 6.383 / SU ML: 0.162 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / ESU R: 0.303 / ESU R Free: 0.224 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.4 Å / Solvent model: BABINET MODEL WITH MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 38.732 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.15→53.45 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.151→2.207 Å / Total num. of bins used: 20 /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller



















 PDBj
PDBj