[English] 日本語
 Yorodumi
Yorodumi- PDB-1hrz: THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIM... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hrz | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIMENSIONAL HETERONUCLEAR-EDITED AND-FILTERED NMR | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA BINDING PROTEIN/DNA / DNA BINDING PROTEIN-DNA complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of male gonad development / Transcriptional regulation of testis differentiation / sex differentiation / male sex determination / Deactivation of the beta-catenin transactivating complex / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / cell differentiation / calmodulin binding ...positive regulation of male gonad development / Transcriptional regulation of testis differentiation / sex differentiation / male sex determination / Deactivation of the beta-catenin transactivating complex / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding / DNA-binding transcription factor activity, RNA polymerase II-specific / cell differentiation / calmodulin binding / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / positive regulation of gene expression / positive regulation of DNA-templated transcription / chromatin / positive regulation of transcription by RNA polymerase II / DNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Clore, G.M. / Werner, M.H. / Huth, J.R. / Gronenborn, A.M. | ||||||
 Citation Citation |  Journal: Cell(Cambridge,Mass.) / Year: 1995 Journal: Cell(Cambridge,Mass.) / Year: 1995Title: Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Authors: Werner, M.H. / Huth, J.R. / Gronenborn, A.M. / Clore, G.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hrz.cif.gz 1hrz.cif.gz | 1.3 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hrz.ent.gz pdb1hrz.ent.gz | 1.1 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hrz.json.gz 1hrz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hr/1hrz https://data.pdbj.org/pub/pdb/validation_reports/hr/1hrz ftp://data.pdbj.org/pub/pdb/validation_reports/hr/1hrz ftp://data.pdbj.org/pub/pdb/validation_reports/hr/1hrz | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 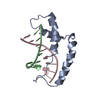
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2404.621 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: DNA chain | Mass: 2448.613 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| #3: Protein | Mass: 9508.031 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: Q05066 Homo sapiens (human) / References: UniProt: Q05066 |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Software ordinal: 1 Details: THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIMENSIONAL HETERONUCLEAR-EDITED AND -FILTERED NMR IS BASED ON 1805 EXPERIMENTAL RESTRAINTS: (A) INTRA-PROTEIN: 290 SEQUENTIAL ...Details: THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIMENSIONAL HETERONUCLEAR-EDITED AND -FILTERED NMR IS BASED ON 1805 EXPERIMENTAL RESTRAINTS: (A) INTRA-PROTEIN: 290 SEQUENTIAL (|I-J|=1), 221 MEDIUM RANGE (1 < |I-J| >=5) AND 107 LONG RANGE (|I-J| >5) INTERRESIDUES. 238 INTRARESIDUE APPROXIMATE INTERPROTON DISTANCE RESTRAINTS; 70 DISTANCE RESTRAINTS FOR 35 HYDROGEN BONDS; 153 TORSION ANGLE (71 PHI, 10 PSI, 56 CHI1 AND 16 CHI2) RESTRAINTS; 56 THREE-BOND HN-HA COUPLING CONSTANT RESTRAINTS; 145 (73 CALPHA AND 72 CBETA) 13C SHIFT RESTRAINTS. (B) INTRA-DNA: 206 INTRARESIDUE, 96 SEQUENTIAL INTRASTRAND, 36 INTERSTRAND INTERPROTON DISTANCE RESTRAINTS; 40 H-BOND RESTRAINTS; 72 TORSION ANGLE RESTRAINTS (FOR ALPHA, BETA, GAMMA, EPSILON AND ZETA BACKBONE TORSION ANGLES. (C) INTERMOLECULAR: 75 INTERPROTON DISTANCE RESTRAINTS THE STRUCTURES IN THIS ENTRY REPRESENT 35 INDIVIDUAL SIMULATED ANNEALING STRUCTURES. THE RESTRAINED REGULARIZED MEAN STRUCTURE CAN BE FOUND IN PDB ENTRY 1HRY. THE LAST COLUMN IN THE INDIVIDUAL SA STRUCTURES HAS NO MEANING. | ||||||||||||
| NMR ensemble | Conformers submitted total number: 35 |
 Movie
Movie Controller
Controller










 PDBj
PDBj

 X-PLOR
X-PLOR