+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1hhx | ||||||
|---|---|---|---|---|---|---|---|
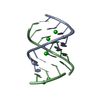
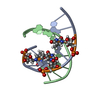
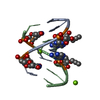
| Title | Solution structure of LNA3:RNA hybrid | ||||||
 Components Components |
| ||||||
 Keywords Keywords | ANTISENSE / LOCKED NUCLEIC ACID / LNA / RNA / HYBRID / RNASE H | ||||||
| Function / homology | DNA/RNA hybrid / RNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Petersen, M. / Bondensgaard, K. / Wengel, J. / Jacobsen, J.P. | ||||||
 Citation Citation |  Journal: J.Am.Chem.Soc. / Year: 2002 Journal: J.Am.Chem.Soc. / Year: 2002Title: Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids Authors: Petersen, M. / Bondensgaard, K. / Wengel, J. / Jacobsen, J.P. #1:  Journal: Chemistry / Year: 2000 Journal: Chemistry / Year: 2000Title: Structural Studies of Lna:RNA Duplexes by NMR: Conformations and Implications for Rnase H Activity. Authors: Bondensgaard, K. / Petersen, M. / Singh, S.K. / Rajwanshi, V.K. / Kumar, R. / Wengel, J. / Jacobsen, J.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1hhx.cif.gz 1hhx.cif.gz | 389.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1hhx.ent.gz pdb1hhx.ent.gz | 320.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1hhx.json.gz 1hhx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hh/1hhx https://data.pdbj.org/pub/pdb/validation_reports/hh/1hhx ftp://data.pdbj.org/pub/pdb/validation_reports/hh/1hhx ftp://data.pdbj.org/pub/pdb/validation_reports/hh/1hhx | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 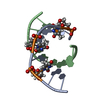
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA/RNA hybrid | Mass: 2814.843 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: RNA chain | Mass: 2855.767 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| Sequence details | CHAIN A RESIDUES 2,5,7 ARE LOCKED NUCLEIC ACIDS OR (1R,3R,4R,7S)-7-HYDROXY-1-HYDROXYMETHYL-3- ...CHAIN A RESIDUES 2,5,7 ARE LOCKED NUCLEIC ACIDS OR (1R,3R,4R,7S)-7-HYDROXY-1-HYDROXYMET |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|---|
| NMR experiment | Type: NOESY |
- Sample preparation
Sample preparation
| Details | Contents: 0.7E-3M LNA1:RNA, 0.1M NACL, 5E-5M EDTA AND 1E-2M PHOSPHATE BUFFER IN 100% D2O |
|---|---|
| Sample conditions | Ionic strength: 0.1 / pH: 7 / Pressure: 1 atm / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model: INOVA / Field strength: 750 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: A TOTAL OF 423 NOE DISTANCE RESTRAINTS WERE OBTAINED FROM A FULL RELAXATION MATRIX ANALYSIS WITH THE PROGRAM RANDMARDI. A FURTHER 58 NOE DISTANCE RESTRAINTS WERE OBTAINED FROM NOESY IN H2O ...Details: A TOTAL OF 423 NOE DISTANCE RESTRAINTS WERE OBTAINED FROM A FULL RELAXATION MATRIX ANALYSIS WITH THE PROGRAM RANDMARDI. A FURTHER 58 NOE DISTANCE RESTRAINTS WERE OBTAINED FROM NOESY IN H2O USING ISPA. 22 WATSON-CRICK BASE PAIRING RESTRAINTS. | |||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST ENERGY / Conformers calculated total number: 40 / Conformers submitted total number: 34 |
 Movie
Movie Controller
Controller







 PDBj
PDBj Amber
Amber