+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fyo | ||||||
|---|---|---|---|---|---|---|---|
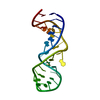
| Title | EUKARYOTIC DECODING REGION A-SITE RNA | ||||||
 Components Components | FRAGMENT OF 18S RIBOSOMAL RNA | ||||||
 Keywords Keywords | RNA / stem-internal loop-stem-tetraloop RNA / G-A base pair / bulged A | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing, molecular dynamics | ||||||
 Authors Authors | Lynch, S.R. / Puglisi, J.D. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2001 Journal: J.Mol.Biol. / Year: 2001Title: Structure of a eukaryotic decoding region A-site RNA. Authors: Lynch, S.R. / Puglisi, J.D. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fyo.cif.gz 1fyo.cif.gz | 411.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fyo.ent.gz pdb1fyo.ent.gz | 350.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fyo.json.gz 1fyo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fy/1fyo https://data.pdbj.org/pub/pdb/validation_reports/fy/1fyo ftp://data.pdbj.org/pub/pdb/validation_reports/fy/1fyo ftp://data.pdbj.org/pub/pdb/validation_reports/fy/1fyo | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 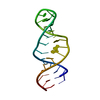
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 8672.185 Da / Num. of mol.: 1 Fragment: 18S RNA - NUCLEOTIDES 1404-1412, 1488-1497 (E. COLI NUMBERS) Mutation: U1410A, A1490U / Source method: obtained synthetically Details: This sequence occurs naturally in Tetrahymena thermophila |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 3 mM decoding site RNA, 13C/15N, 10 mM phosphate pH 6.3, 90% H20/10% D20, 100% D20 Solvent system: 90% H2O/10% D20; 100% D2O |
|---|---|
| Sample conditions | Ionic strength: 10 mM / pH: 6.3 / Pressure: 1 atm / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing, molecular dynamics / Software ordinal: 1 Details: 682 NOEs, 129 dihedral constraints, 36 Hydrogen bonds | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations,structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 25 |
 Movie
Movie Controller
Controller










 PDBj
PDBj





























 X-PLOR
X-PLOR