[English] 日本語
 Yorodumi
Yorodumi- PDB-1fm1: SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1fm1 | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | ||||||
 Components Components | COLLAGENASE-3 | ||||||
 Keywords Keywords | HYDROLASE / Matrix Metalloproteinase / Hydroxamic acid / Human Collagenase-3 / MMP-13 | ||||||
| Function / homology |  Function and homology information Function and homology informationgrowth plate cartilage development / RUNX2 regulates genes involved in cell migration / endochondral ossification / bone morphogenesis / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / Assembly of collagen fibrils and other multimeric structures / Activation of Matrix Metalloproteinases / bone mineralization / Collagen degradation / response to amyloid-beta ...growth plate cartilage development / RUNX2 regulates genes involved in cell migration / endochondral ossification / bone morphogenesis / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / Assembly of collagen fibrils and other multimeric structures / Activation of Matrix Metalloproteinases / bone mineralization / Collagen degradation / response to amyloid-beta / collagen catabolic process / extracellular matrix disassembly / collagen binding / Degradation of the extracellular matrix / extracellular matrix organization / metalloendopeptidase activity / extracellular matrix / endopeptidase activity / serine-type endopeptidase activity / calcium ion binding / proteolysis / extracellular space / extracellular region / zinc ion binding Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / distance geometry simulated annealing | ||||||
 Authors Authors | Moy, F.J. / Chanda, P.K. / Chen, J.M. / Cosmi, S. / Edris, W. / Levin, J.I. / Powers, R. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2000 Journal: J.Mol.Biol. / Year: 2000Title: High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor. Authors: Moy, F.J. / Chanda, P.K. / Chen, J.M. / Cosmi, S. / Edris, W. / Levin, J.I. / Powers, R. #1:  Journal: J.Biomol.NMR / Year: 2000 Journal: J.Biomol.NMR / Year: 2000Title: 1H, 15N, 13C, and 13CO Assignments and Secondary Structure Determination of Collagenase-3 (MMP-13) Complexed with a Hydroxamic Acid Inhibitor Authors: Moy, F.J. / Chanda, P.K. / Cosmi, S. / Edris, W. / Levin, J.I. / Powers, R. | ||||||
| History |
|
- Structure visualization
Structure visualization
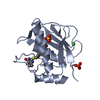
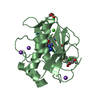
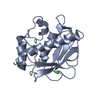
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1fm1.cif.gz 1fm1.cif.gz | 1.4 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1fm1.ent.gz pdb1fm1.ent.gz | 1.2 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1fm1.json.gz 1fm1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fm/1fm1 https://data.pdbj.org/pub/pdb/validation_reports/fm/1fm1 ftp://data.pdbj.org/pub/pdb/validation_reports/fm/1fm1 ftp://data.pdbj.org/pub/pdb/validation_reports/fm/1fm1 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 18607.824 Da / Num. of mol.: 1 / Fragment: CATALYTIC FRAGMENT Source method: isolated from a genetically manipulated source Details: HYDROXAMIC ACID INHIBITOR COMPLEX / Source: (gene. exp.)  Homo sapiens (human) / Plasmid: PPROMMP-13 / Production host: Homo sapiens (human) / Plasmid: PPROMMP-13 / Production host:  References: UniProt: P45452, Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases | ||||
|---|---|---|---|---|---|
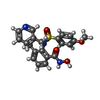
| #2: Chemical | | #3: Chemical | ChemComp-CA / | #4: Chemical | ChemComp-WAY / | |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: The structure was determined using triple-resonance and isotope filtered NMR spectroscopy. |
- Sample preparation
Sample preparation
| Details |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Ionic strength: 10 mM deuterated Tris-Base, 100 mM NaCl, 5 mM CaCl2, 0.1 mM ZnCl2, 2 mM NaN3, 10 mM deuterated DTT pH: 6.5 / Pressure: ambient / Temperature: 308 K | ||||||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AMX 2 / Manufacturer: Bruker / Model: AMX 2 / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: distance geometry simulated annealing / Software ordinal: 1 Details: structures calculated were based on 3280 experimental NMR restraints, consisting of 2415 approximate interproton distance restraints, 47 distance restraints between MMP-13 and WAY-151693, 5 ...Details: structures calculated were based on 3280 experimental NMR restraints, consisting of 2415 approximate interproton distance restraints, 47 distance restraints between MMP-13 and WAY-151693, 5 intramolecular distance restraints for WAY-151693, 88 distance restraints for 44 backbone hydrogen bonds, 391 torsion angle restraints, 103 3JNHa restraints 123 Ca restraints and 108 Cb restraints. The structure was also refined using a conformational database. | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with acceptable covalent geometry,structures with favorable non-bond energy,structures with the least restraint violations,structures with the lowest energy,target function Conformers calculated total number: 100 / Conformers submitted total number: 30 |
 Movie
Movie Controller
Controller










 PDBj
PDBj









 NMRPipe
NMRPipe