[English] 日本語
 Yorodumi
Yorodumi- PDB-4jqg: Crystal structure of an inactive mutant of MMP-9 catalytic domain... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4jqg | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of an inactive mutant of MMP-9 catalytic domain in complex with a fluorogenic synthetic peptidic substrate with a fluorine atom. | ||||||
 Components Components |
| ||||||
 Keywords Keywords | HYDROLASE/substrate / halogen-water-hydrogen bridge / Zincin-like / Gelatinase / Collagenase / Catalytic Domain / HYDROLASE-substrate complex | ||||||
| Function / homology |  Function and homology information Function and homology informationgelatinase B / negative regulation of epithelial cell differentiation involved in kidney development / : / : / cellular response to UV-A / regulation of neuroinflammatory response / positive regulation of keratinocyte migration / Assembly of collagen fibrils and other multimeric structures / positive regulation of DNA binding / Activation of Matrix Metalloproteinases ...gelatinase B / negative regulation of epithelial cell differentiation involved in kidney development / : / : / cellular response to UV-A / regulation of neuroinflammatory response / positive regulation of keratinocyte migration / Assembly of collagen fibrils and other multimeric structures / positive regulation of DNA binding / Activation of Matrix Metalloproteinases / negative regulation of intrinsic apoptotic signaling pathway / endodermal cell differentiation / Collagen degradation / positive regulation of release of cytochrome c from mitochondria / response to amyloid-beta / collagen catabolic process / macrophage differentiation / extracellular matrix disassembly / EPH-ephrin mediated repulsion of cells / ephrin receptor signaling pathway / positive regulation of epidermal growth factor receptor signaling pathway / collagen binding / positive regulation of vascular associated smooth muscle cell proliferation / Degradation of the extracellular matrix / extracellular matrix organization / embryo implantation / skeletal system development / Signaling by SCF-KIT / metalloendopeptidase activity / : / positive regulation of protein phosphorylation / metallopeptidase activity / tertiary granule lumen / cell migration / peptidase activity / cellular response to lipopolysaccharide / Interleukin-4 and Interleukin-13 signaling / endopeptidase activity / ficolin-1-rich granule lumen / Extra-nuclear estrogen signaling / positive regulation of apoptotic process / serine-type endopeptidase activity / apoptotic process / Neutrophil degranulation / negative regulation of apoptotic process / proteolysis / extracellular space / extracellular exosome / extracellular region / zinc ion binding / identical protein binding Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.849 Å MOLECULAR REPLACEMENT / Resolution: 1.849 Å | ||||||
 Authors Authors | Stura, E.A. / Vera, L. / Cassar-Lajeunesse, E. / Tranchant, I. / Amoura, M. / Dive, V. | ||||||
 Citation Citation |  Journal: Chem.Biol. / Year: 2014 Journal: Chem.Biol. / Year: 2014Title: Halogen Bonding Controls Selectivity of FRET Substrate Probes for MMP-9. Authors: Tranchant, I. / Vera, L. / Czarny, B. / Amoura, M. / Cassar, E. / Beau, F. / Stura, E.A. / Dive, V. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4jqg.cif.gz 4jqg.cif.gz | 102.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4jqg.ent.gz pdb4jqg.ent.gz | 75.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4jqg.json.gz 4jqg.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jq/4jqg https://data.pdbj.org/pub/pdb/validation_reports/jq/4jqg ftp://data.pdbj.org/pub/pdb/validation_reports/jq/4jqg ftp://data.pdbj.org/pub/pdb/validation_reports/jq/4jqg | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4jijSC  4jxa S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein/peptide / Protein , 2 types, 4 molecules PQAB
| #1: Protein/peptide | Mass: 1145.157 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: fluorogenic synthetic L-peptide substrate with lower catalytic efficiency with MMP-9 than the iodine containing substrate PDB code 4JIJ. #2: Protein | Mass: 18280.283 Da / Num. of mol.: 2 / Mutation: E402A Source method: isolated from a genetically manipulated source Details: Catalytic domain construct: 110-216 and 392-444 Mutagenesis: Glu402Ala Structur e: renumbered omitting missing domain. Source: (gene. exp.)  Homo sapiens (human) / Gene: CLG4B, MMP9 / Plasmid: pET-14b / Production host: Homo sapiens (human) / Gene: CLG4B, MMP9 / Plasmid: pET-14b / Production host:  |
|---|
-Non-polymers , 9 types, 453 molecules 





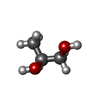










| #3: Chemical | | #4: Chemical | ChemComp-ZN / #5: Chemical | #6: Chemical | #7: Chemical | ChemComp-EDO / #8: Chemical | ChemComp-GOL / | #9: Chemical | #10: Chemical | #11: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.16 Å3/Da / Density % sol: 43.18 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 6 Details: Protein: hMMP9 E402A at 420 microM + AHA 120mM 0.12 M acetohydroxamic acid and 0.5 microL IT34-F at 10 milli-M. Reservoir: 10% PEG 20K, 0.1 M MMT (L-malic acid, MES, Tris) 75% acid/25% ...Details: Protein: hMMP9 E402A at 420 microM + AHA 120mM 0.12 M acetohydroxamic acid and 0.5 microL IT34-F at 10 milli-M. Reservoir: 10% PEG 20K, 0.1 M MMT (L-malic acid, MES, Tris) 75% acid/25% basic, 0.5 M NaCl, 0.02 SrCl2, 10% glycerol. Cryoprotectant: 10% Di-ethylene glycol, 5% glycerol, 10% propanediol, 5% dioxane, .8 M Li formate, 9% PEG 10,000 0.1M (MMT 75/25), pH 6.0, VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-1 / Wavelength: 0.972981 Å / Beamline: ID23-1 / Wavelength: 0.972981 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Mar 7, 2013 / Details: mirrors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Channel cut Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.972981 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.849→50 Å / Num. all: 29876 / Num. obs: 29657 / % possible obs: 99.3 % / Observed criterion σ(F): 0 / Observed criterion σ(I): -3 / Redundancy: 8.46 % / Biso Wilson estimate: 25.048 Å2 / Rmerge(I) obs: 0.343 / Rsym value: 0.325 / Net I/σ(I): 5.29 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4JIJ Resolution: 1.849→40.524 Å / SU ML: 0.22 / Isotropic thermal model: Isotropic / Cross valid method: THROUGHOUT / σ(F): 1.99 / σ(I): -3 / Phase error: 26.08 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 25.048 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.849→40.524 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION
|
 Movie
Movie Controller
Controller














 PDBj
PDBj



















