[English] 日本語
 Yorodumi
Yorodumi- PDB-1elf: NATURE OF THE INACTIVATION OF ELASTASE BY N-PEPTIDYL-O-AROYL HYDR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1elf | ||||||
|---|---|---|---|---|---|---|---|
| Title | NATURE OF THE INACTIVATION OF ELASTASE BY N-PEPTIDYL-O-AROYL HYDROXYLAMINE AS A FUNCTION OF PH | ||||||
 Components Components | PORCINE PANCREATIC ELASTASE | ||||||
 Keywords Keywords | COMPLEX (HYDROLASE/INHIBITOR) / COMPLEX (HYDROLASE-INHIBITOR) / COMPLEX (HYDROLASE-INHIBITOR) complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpancreatic elastase / serine-type endopeptidase activity / proteolysis / extracellular space / metal ion binding Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.7 Å X-RAY DIFFRACTION / Resolution: 1.7 Å | ||||||
 Authors Authors | Ding, X. / Rasmussen, B. / Demuth, H.-U. / Ringe, D. / Steinmetz, A.C.U. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1995 Journal: Biochemistry / Year: 1995Title: Nature of the inactivation of elastase by N-peptidyl-O-aroyl hydroxylamine as a function of pH. Authors: Ding, X. / Rasmussen, B.F. / Demuth, H.U. / Ringe, D. / Steinmetz, A.C. #1:  Journal: Biochemistry / Year: 1994 Journal: Biochemistry / Year: 1994Title: Direct Structural Observation of an Acyl-Enzyme Intermediate in the Hydrolysis of an Ester Substrate by Elastase Authors: Ding, X. / Rasmussen, B.F. / Petsko, G.A. / Ringe, D. #2:  Journal: J.Enzyme Inhib. / Year: 1991 Journal: J.Enzyme Inhib. / Year: 1991Title: Inhibition of Proteases with Enkephalin-Analogue Inhibitors Authors: Demuth, H.-U. / Silberring, J. / Nyberg, F. #3:  Journal: J.Am.Chem.Soc. / Year: 1991 Journal: J.Am.Chem.Soc. / Year: 1991Title: Competing Redox and Inactivation Processes in the Inhibition of Cysteine Proteinases by Peptidyl O-Acylhydroxamates. 13C and 15N NMR Evidence for a Novel Sulfenamide Enzyme Adduct Authors: Robinson, V.J. / Coles, P.J. / Smith, R.A. / Krantz, A. #4:  Journal: J.Org.Chem. / Year: 1989 Journal: J.Org.Chem. / Year: 1989Title: N-O Bond Fission as the Rate-Determining Step in the Aqueous Conversion of N-Peptidyl-O-(P-Nitrobenzoyl)Hydroxylamines to P-Nitrobenzoic Acid and Peptidylhydroxamic Acids Authors: Demuth, H.-U. / Fischer, G. / Barth, A. / Schowen, R.L. #5:  Journal: J.Am.Chem.Soc. / Year: 1989 Journal: J.Am.Chem.Soc. / Year: 1989Title: Crystal Structure of the Covalent Complex Formed by a Peptidyl Difluoro Keto Amide with Porcine Pancreatic Elastase at 1.78 Angstroms Resolution Authors: Takahashi, L.H. / Radhakrishnan, R. / Rosenfield Junior, R.E. / Meyer Junior, E.F. / Trainor, D.A. | ||||||
| History |
| ||||||
| Remark 700 | SHEET THE TWO SEVEN-STRANDED SHEETS IN THIS STRUCTURE ARE REALLY SIX-STRANDED BETA BARRELS. THIS IS ...SHEET THE TWO SEVEN-STRANDED SHEETS IN THIS STRUCTURE ARE REALLY SIX-STRANDED BETA BARRELS. THIS IS DENOTED BY THE FIRST STRAND RECURRING AS THE LAST STRAND. |
- Structure visualization
Structure visualization
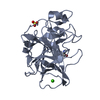
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1elf.cif.gz 1elf.cif.gz | 64.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1elf.ent.gz pdb1elf.ent.gz | 46.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1elf.json.gz 1elf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/el/1elf https://data.pdbj.org/pub/pdb/validation_reports/el/1elf ftp://data.pdbj.org/pub/pdb/validation_reports/el/1elf ftp://data.pdbj.org/pub/pdb/validation_reports/el/1elf | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 25928.031 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  | ||||||
|---|---|---|---|---|---|---|---|
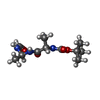
| #2: Chemical | ChemComp-CA / | ||||||
| #3: Chemical | | #4: Chemical | ChemComp-BAF / ( | #5: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.2 Å3/Da / Density % sol: 44.21 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS pH: 5 / Method: vapor diffusion, sitting drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| Reflection | Resolution: 1.7→10 Å / Num. obs: 21705 / % possible obs: 84 % / Observed criterion σ(I): 1 |
| Reflection | *PLUS Highest resolution: 1.68 Å / Lowest resolution: 22.36 Å / Num. obs: 23713 / % possible obs: 91 % / Observed criterion σ(I): 0 / Rmerge(I) obs: 0.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Rfactor Rwork: 0.18 / Rfactor obs: 0.18 / Highest resolution: 1.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 1.7 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 10 Å / Num. reflection all: 21705 / σ(I): 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller











 PDBj
PDBj