+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1e7r | ||||||
|---|---|---|---|---|---|---|---|
| Title | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | ||||||
 Components Components | GDP-FUCOSE SYNTHETASE | ||||||
 Keywords Keywords | ISOMERASE / EPIMERASE / REDUCTASE / SDR / RED | ||||||
| Function / homology |  Function and homology information Function and homology informationGDP-L-fucose synthase / GDP-L-fucose synthase activity / colanic acid biosynthetic process / 'de novo' GDP-L-fucose biosynthetic process / NADP+ binding / isomerase activity / protein homodimerization activity / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.6 Å MOLECULAR REPLACEMENT / Resolution: 1.6 Å | ||||||
 Authors Authors | Rosano, C. / Izzo, G. / Bolognesi, M. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2000 Journal: J.Mol.Biol. / Year: 2000Title: Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants Authors: Rosano, C. / Bisso, A. / Izzo, G. / Tonetti, M. / Sturla, L. / De Flora, A. / Bolognesi, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1e7r.cif.gz 1e7r.cif.gz | 90 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1e7r.ent.gz pdb1e7r.ent.gz | 65.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1e7r.json.gz 1e7r.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e7/1e7r https://data.pdbj.org/pub/pdb/validation_reports/e7/1e7r ftp://data.pdbj.org/pub/pdb/validation_reports/e7/1e7r ftp://data.pdbj.org/pub/pdb/validation_reports/e7/1e7r | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1e6uC  1e7qC  1e7sC  1bwsS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 36126.016 Da / Num. of mol.: 1 / Mutation: YES Source method: isolated from a genetically manipulated source Details: UNKNOWN MOLECULE LABELED AS ACETYLPHOSPHATE / Source: (gene. exp.)   References: UniProt: P32055, Isomerases; Racemases and epimerases; Acting on carbohydrates and derivatives |
|---|
-Non-polymers , 5 types, 353 molecules 
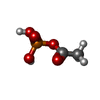







| #2: Chemical | ChemComp-NAP / | ||||
|---|---|---|---|---|---|
| #3: Chemical | ChemComp-UVW / | ||||
| #4: Chemical | | #5: Chemical | #6: Water | ChemComp-HOH / | |
-Details
| Compound details | CHAIN A ENGINEERED MUTATION TYR136GLU NADP-DEPENDENT CONVERSION OF GDP-4-DEHYDRO-6-DEOXY-D-MANNOSE ...CHAIN A ENGINEERED |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.2 Å3/Da / Density % sol: 61.53 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 294 K / pH: 6.5 Details: 1.5 M LITHIUM SULPHATE, PH 6.5 0.1M TRIS BUFFER 21C | ||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 21 ℃ / Method: unknown / PH range low: 7.8 / PH range high: 6.5 | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  EMBL/DESY, HAMBURG EMBL/DESY, HAMBURG  / Beamline: BW7A / Wavelength: 0.844 / Beamline: BW7A / Wavelength: 0.844 |
| Detector | Type: MARRESEARCH |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.844 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→100 Å / Num. obs: 61196 / % possible obs: 99.3 % / Redundancy: 6.2 % / Rmerge(I) obs: 0.043 / Net I/σ(I): 9.65 |
| Reflection shell | Resolution: 1.6→1.63 Å / Mean I/σ(I) obs: 2.4 / % possible all: 99.1 |
| Reflection shell | *PLUS % possible obs: 99.1 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1BWS Resolution: 1.6→10 Å / σ(F): 0 / ESU R: 0.069 / ESU R Free: 0.071
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller
















 PDBj
PDBj



