+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1dy4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
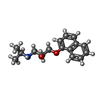
| Title | CBH1 IN COMPLEX WITH S-PROPRANOLOL | |||||||||
 Components Components | EXOGLUCANASE 1 | |||||||||
 Keywords Keywords | HYDROLASE / HYDROLASE(O-GLYCOSYL) / CELLULOSE DEAGRADATION / CHIRAL SEPARATION / GLYCOSIDASE / GLYCOPROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellulose 1,4-beta-cellobiosidase (non-reducing end) / cellulose 1,4-beta-cellobiosidase activity / cellulose binding / cellulose catabolic process / extracellular region Similarity search - Function | |||||||||
| Biological species |  TRICHODERMA REESEI (fungus) TRICHODERMA REESEI (fungus) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.9 Å MOLECULAR REPLACEMENT / Resolution: 1.9 Å | |||||||||
 Authors Authors | Stahlberg, J. / Henriksson, H. / Divne, C. / Isaksson, R. / Pettersson, G. / Johansson, G. / Jones, T.A. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2001 Journal: J.Mol.Biol. / Year: 2001Title: Structural Basis for Enantiomer Binding and Separation of a Common Beta-Blocker: Crystal Structure of Cellobiohydrolase Cel7A with Bound (S)-Propranolol at 1.9 A Resolution Authors: Stahlberg, J. / Henriksson, H. / Divne, C. / Isaksson, R. / Pettersson, G. / Johansson, G. / Jones, T.A. #1:  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: High-Resolution Crystal Structures Reveal How a Cellulose Chain is Bound in the 50A Long Tunnel of Cellobiohydrolase I from Trichoderma Reesei Authors: Divne, C. / Stahlberg, J. / Teeri, T.T. / Jones, T.A. #2:  Journal: Science / Year: 1994 Journal: Science / Year: 1994Title: The Three-Dimensional Crystal Structure of the Catalytic Core of Cellobiohydrolase I from Trichoderma Reesei Authors: Divne, C. / Stahlberg, J. / Reinikainen, T. / Ruohonen, L. / Pettersson, G. / Knowles, J.K. / Teeri, T.T. / Jones, T.A. | |||||||||
| History |
| |||||||||
| Remark 700 | SHEET DETERMINATION METHOD: THERE IS A BIFURCATED SHEET IN THIS STRUCTURE. THIS IS REPRESENTED BY ... SHEET DETERMINATION METHOD: THERE IS A BIFURCATED SHEET IN THIS STRUCTURE. THIS IS REPRESENTED BY TWO SHEETS WHICH HAVE ONE OR MORE IDENTICAL STRANDS. SHEETS A AND A1 REPRESENT ONE BIFURCATED SHEET. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1dy4.cif.gz 1dy4.cif.gz | 103.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1dy4.ent.gz pdb1dy4.ent.gz | 78.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1dy4.json.gz 1dy4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/dy/1dy4 https://data.pdbj.org/pub/pdb/validation_reports/dy/1dy4 ftp://data.pdbj.org/pub/pdb/validation_reports/dy/1dy4 ftp://data.pdbj.org/pub/pdb/validation_reports/dy/1dy4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5celS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 46010.703 Da / Num. of mol.: 1 / Fragment: CATALYTIC DOMAIN, RESIDUES 18-451 / Source method: isolated from a natural source / Source: (natural)  TRICHODERMA REESEI (fungus) / Strain: QM9414 TRICHODERMA REESEI (fungus) / Strain: QM9414References: UniProt: P62694, cellulose 1,4-beta-cellobiosidase (non-reducing end) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #2: Sugar | | #3: Chemical | ChemComp-SNP / | #4: Chemical | #5: Water | ChemComp-HOH / | Compound details | THE I222 CRYSTAL FORM DESCRIBED HERE REPRESENTS THE HIGHER SYMMETRY EQUIVALENT TO THE PSEUDO-I222 ...THE I222 CRYSTAL FORM DESCRIBED HERE REPRESENTS | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.1 Å3/Da / Density % sol: 41 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Method: vapor diffusion, hanging drop / pH: 7 Details: HANGING DROPS. EQUAL VOLUMES OF 9 MG/ML PROTEIN/7.5 MM S-PROPRANOLOL AND RESERVOIR SOLUTION CONTAINING 0.1 M MES (PH 7.0), 24% (W/V) MONOMETHYL ETHER PEG 5000, 15% GLYCEROL AND 10 MM COCL2. | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: R-AXIS IIC / Detector: IMAGE PLATE / Details: MIRRORS |
| Radiation | Monochromator: GRAPHITE(002) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→33.6 Å / Num. obs: 30518 / % possible obs: 100 % / Redundancy: 12.3 % / Rmerge(I) obs: 0.043 / Net I/σ(I): 32.6 |
| Reflection shell | Resolution: 1.9→1.97 Å / Rmerge(I) obs: 0.095 / Mean I/σ(I) obs: 12.3 / % possible all: 100 |
| Reflection | *PLUS Num. measured all: 375375 |
| Reflection shell | *PLUS % possible obs: 100 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 5CEL Resolution: 1.9→30 Å / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→30 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file | Serial no: 1 / Param file: PARHCSDX.PRO / Topol file: TOPHCSDX.PRO | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Version: 1 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Num. reflection obs: 28512 / Rfactor obs: 0.197 / Rfactor Rfree: 0.224 / Rfactor Rwork: 0.197 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller
























 PDBj
PDBj