[English] 日本語
 Yorodumi
Yorodumi- PDB-1df7: DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED W... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1df7 | ||||||
|---|---|---|---|---|---|---|---|
| Title | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND METHOTREXATE | ||||||
 Components Components | DIHYDROFOLATE REDUCTASE | ||||||
 Keywords Keywords | OXIDOREDUCTASE / DIHYDROFOLATE REDUCTASE / STRUCTURE-BASED INHIBITOR DESIGN / FOLATEANALOGS / ROSSMANN FOLD / NICOTINAMIDE ADENINE DINUCLEOTIDE / METHOTREXATE / TUBERCULOSIS | ||||||
| Function / homology |  Function and homology information Function and homology informationglycine biosynthetic process / NADP+ binding / dihydrofolate metabolic process / dihydrofolate reductase / dihydrofolate reductase activity / folic acid metabolic process / tetrahydrofolate biosynthetic process / one-carbon metabolic process / NADP binding / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.7 Å SYNCHROTRON / Resolution: 1.7 Å | ||||||
 Authors Authors | Li, R. / Sirawaraporn, R. / Chitnumsub, P. / Sirawaraporn, W. / Wooden, J. / Athappilly, F. / Turley, S. / Hol, W.G. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2000 Journal: J.Mol.Biol. / Year: 2000Title: Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs. Authors: Li, R. / Sirawaraporn, R. / Chitnumsub, P. / Sirawaraporn, W. / Wooden, J. / Athappilly, F. / Turley, S. / Hol, W.G. #1:  Journal: Biochemistry / Year: 1997 Journal: Biochemistry / Year: 1997Title: Comparison of Two Independent Crystal Structures of Human Dihydrofolate Reductase Ternary Complexes Reduced with Nicotinamide Adenine Dinucleotide Phosphate and the Very Tight-Binding Inhibitor PT523 Authors: Cody, V. / Galitsky, N. / Luft, J.R. / Pangborn, W. / Rosowsky, A. / Blakley, R.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1df7.cif.gz 1df7.cif.gz | 52.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1df7.ent.gz pdb1df7.ent.gz | 36.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1df7.json.gz 1df7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/df/1df7 https://data.pdbj.org/pub/pdb/validation_reports/df/1df7 ftp://data.pdbj.org/pub/pdb/validation_reports/df/1df7 ftp://data.pdbj.org/pub/pdb/validation_reports/df/1df7 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 17660.992 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  References: UniProt: P0A546, UniProt: P9WNX1*PLUS, dihydrofolate reductase |
|---|
-Non-polymers , 5 types, 202 molecules 

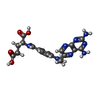






| #2: Chemical | ChemComp-SO4 / | ||
|---|---|---|---|
| #3: Chemical | ChemComp-NDP / | ||
| #4: Chemical | ChemComp-MTX / | ||
| #5: Chemical | ChemComp-GOL / #6: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.05 Å3/Da / Density % sol: 59.64 % | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion / pH: 4.5 Details: AMMONIUM SULFATE, SODIUM ACETATE, GLYCEROL, NADPH, METHOTREXATE, POTASSIUM PHOSPHATE, DTT, POTASSIUM CHLORIDE, pH 4.5, VAPOR DIFFUSION, temperature 277K | ||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 1.0188 / Beamline: 19-ID / Wavelength: 1.0188 |
| Detector | Type: SBC-2 / Detector: CCD / Date: Sep 17, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.0188 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→50 Å / Num. all: 23394 / Num. obs: 109952 / % possible obs: 99.7 % / Redundancy: 4.7 % / Rmerge(I) obs: 0.042 / Net I/σ(I): 27 |
| Reflection shell | Resolution: 1.7→1.78 Å / Redundancy: 4.7 % / Rmerge(I) obs: 0.263 / % possible all: 99.7 |
| Reflection | *PLUS Num. obs: 23394 / Num. measured all: 109952 |
| Reflection shell | *PLUS % possible obs: 99.7 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.7→50 Å / σ(F): 1 / Stereochemistry target values: ENGH & HUBER
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→50 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS % reflection Rfree: 10 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller











 PDBj
PDBj





