[English] 日本語
 Yorodumi
Yorodumi- PDB-1d38: INFLUENCE OF AGLYCONE MODIFICATIONS ON THE BINDING OF ANTHRACYCLI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1d38 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
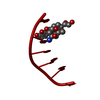
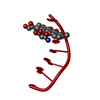
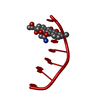
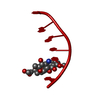
| Title | INFLUENCE OF AGLYCONE MODIFICATIONS ON THE BINDING OF ANTHRACYCLINE DRUGS TO DNA: THE MOLECULAR STRUCTURE OF IDARUBICIN AND 4-O-DEMETHYL-11-DEOXYDOXORUBICIN COMPLEXED TO D(CGATCG) | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords KeywordsDNA / RIGHT HANDED DNA / DOUBLE HELIX / COMPLEXED WITH DRUG | Function / homology | IDARUBICIN / DNA |  Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / Resolution: 1.7 Å X-RAY DIFFRACTION / Resolution: 1.7 Å  Authors AuthorsGao, Y.-G. / Wang, A.H.-J. |  Citation Citation Journal: Anti-Cancer Drug Des. / Year: 1991 Journal: Anti-Cancer Drug Des. / Year: 1991Title: Influence of aglycone modifications on the binding of anthracycline drugs to DNA: the molecular structure of idarubicin and 4-O-demethyl-11-deoxydoxorubicin complexed to d(CGATCG). Authors: Gao, Y.G. / Wang, A.H. #1:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1991 Journal: Proc.Natl.Acad.Sci.USA / Year: 1991Title: Facile Formation of Crosslinked Adduct between DNA and the Daunorubicin Derivative MAR70 Mediated by Formaldehyde: Molecular Structure of the MAR70-d(CGTnACG) Covalent Adduct Authors: Gao, Y.-G. / Liaw, Y.-C. / Li, Y.-K. / Van Der Marel, G.A. / Van Boom, J.H. / Wang, A.H.-J. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1d38.cif.gz 1d38.cif.gz | 16.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1d38.ent.gz pdb1d38.ent.gz | 9.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1d38.json.gz 1d38.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d3/1d38 https://data.pdbj.org/pub/pdb/validation_reports/d3/1d38 ftp://data.pdbj.org/pub/pdb/validation_reports/d3/1d38 ftp://data.pdbj.org/pub/pdb/validation_reports/d3/1d38 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 1809.217 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: Chemical | ChemComp-DM5 / |
| #3: Chemical | ChemComp-MG / |
| #4: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.93 Å3/Da / Density % sol: 58.09 % | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion / pH: 6 / Details: pH 6.00, VAPOR DIFFUSION, temperature 298.00K | ||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 6 / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 298 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 |
| Detector | Type: RIGAKU AFC-5R / Detector: DIFFRACTOMETER |
| Radiation | Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Highest resolution: 1.7 Å / Num. obs: 1724 / Observed criterion σ(F): 2 |
| Reflection | *PLUS Highest resolution: 1.7 Å |
- Processing
Processing
| Software | Name: NUCLSQ / Classification: refinement | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Highest resolution: 1.7 Å / σ(F): 2 /
| ||||||||||||
| Refine Biso |
| ||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 1.7 Å
| ||||||||||||
| Refinement | *PLUS Highest resolution: 1.7 Å / Rfactor obs: 0.188 / Num. reflection obs: 1724 | ||||||||||||
| Solvent computation | *PLUS | ||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller







 PDBj
PDBj






