+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1d1j | ||||||
|---|---|---|---|---|---|---|---|
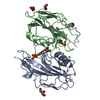
| Title | CRYSTAL STRUCTURE OF HUMAN PROFILIN II | ||||||
 Components Components | PROFILIN II | ||||||
 Keywords Keywords | CONTRACTILE PROTEIN / ACIDIC PROFILIN ISOFORM / ACTIN-BINDING PROTEIN / POLY-L-PROLINE BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationpresynaptic actin cytoskeleton organization / negative regulation of ruffle assembly / negative regulation of epithelial cell migration / modification of postsynaptic actin cytoskeleton / positive regulation of actin filament bundle assembly / regulation of actin filament polymerization / Signaling by ROBO receptors / regulation of synaptic vesicle exocytosis / positive regulation of actin filament polymerization / actin monomer binding ...presynaptic actin cytoskeleton organization / negative regulation of ruffle assembly / negative regulation of epithelial cell migration / modification of postsynaptic actin cytoskeleton / positive regulation of actin filament bundle assembly / regulation of actin filament polymerization / Signaling by ROBO receptors / regulation of synaptic vesicle exocytosis / positive regulation of actin filament polymerization / actin monomer binding / positive regulation of stress fiber assembly / phosphatidylinositol-4,5-bisphosphate binding / presynaptic modulation of chemical synaptic transmission / RHO GTPases Activate Formins / Schaffer collateral - CA1 synapse / presynapse / actin binding / actin cytoskeleton organization / cytoskeleton / postsynapse / protein stabilization / glutamatergic synapse / ATP hydrolysis activity / extracellular exosome / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.2 Å SYNCHROTRON / Resolution: 2.2 Å | ||||||
 Authors Authors | Nodelman, I.M. / Bowman, G.D. / Lindberg, U. / Schutt, C.E. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1999 Journal: J.Mol.Biol. / Year: 1999Title: X-ray structure determination of human profilin II: A comparative structural analysis of human profilins. Authors: Nodelman, I.M. / Bowman, G.D. / Lindberg, U. / Schutt, C.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1d1j.cif.gz 1d1j.cif.gz | 119 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1d1j.ent.gz pdb1d1j.ent.gz | 93.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1d1j.json.gz 1d1j.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d1/1d1j https://data.pdbj.org/pub/pdb/validation_reports/d1/1d1j ftp://data.pdbj.org/pub/pdb/validation_reports/d1/1d1j ftp://data.pdbj.org/pub/pdb/validation_reports/d1/1d1j | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14973.104 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Organ: BRAIN / Plasmid: PET3D / Production host: Homo sapiens (human) / Organ: BRAIN / Plasmid: PET3D / Production host:  #2: Chemical | ChemComp-SO4 / #3: Chemical | ChemComp-PG6 / | #4: Chemical | ChemComp-PG5 / | #5: Water | ChemComp-HOH / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.36 Å3/Da / Density % sol: 47.81 % | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7.3 Details: MAGNESIUM SULFATE, PEG400, pH 7.3, VAPOR DIFFUSION, HANGING DROP, temperature 277K | ||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 4 ℃ | ||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X9B / Wavelength: 0.919 / Beamline: X9B / Wavelength: 0.919 |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Apr 21, 1999 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.919 Å / Relative weight: 1 |
| Reflection | Resolution: 2.2→30 Å / Num. all: 28050 / Num. obs: 28050 / % possible obs: 97.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 2.7 % / Biso Wilson estimate: 17.4 Å2 / Rmerge(I) obs: 0.089 / Net I/σ(I): 10.3 |
| Reflection shell | Resolution: 2.2→2.28 Å / Redundancy: 2.3 % / Rmerge(I) obs: 0.185 / % possible all: 82.8 |
| Reflection | *PLUS Lowest resolution: 50 Å / % possible obs: 96.9 % / Num. measured all: 430124 |
| Reflection shell | *PLUS % possible obs: 85.3 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.2→20 Å / σ(F): 2 / σ(I): 0 / Stereochemistry target values: ENGH & HUBER
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS % reflection Rfree: 5.3 % / Rfactor Rfree: 0.2678 / Rfactor Rwork: 0.2151 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS Biso mean: 23 Å2 |
 Movie
Movie Controller
Controller












 PDBj
PDBj