+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1cyi | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CYTOCHROME C6 | |||||||||
 Components Components | CYTOCHROME C6 | |||||||||
 Keywords Keywords | ELECTRON TRANSPORT PROTEIN (CYTOCHROME) / PHOTOSYNTHESIS / CHLAMYDOMONAS | |||||||||
| Function / homology |  Function and homology information Function and homology informationchloroplast thylakoid lumen / photosynthesis / electron transfer activity / iron ion binding / heme binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.9 Å X-RAY DIFFRACTION / Resolution: 1.9 Å | |||||||||
 Authors Authors | Kerfeld, C.A. / Yeates, T.O. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1995 Journal: J.Mol.Biol. / Year: 1995Title: The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization. Authors: Kerfeld, C.A. / Anwar, H.P. / Interrante, R. / Merchant, S. / Yeates, T.O. #1:  Journal: J.Biol.Chem. / Year: 1991 Journal: J.Biol.Chem. / Year: 1991Title: Isolation and Structural Characterization of the Chlamydomonas Reinhardtii Gene for Cytochrome C6 Authors: Hill, K.L. / Li, H.H. / Singer, J. / Merchant, S. | |||||||||
| History |
| |||||||||
| Remark 700 | SHEET A SHORT TWO-STRANDED ANTI-PARALLEL BETA-SHEET IS FORMED BY RESIDUES 54 - 63, ENCLOSING A TYPE ...SHEET A SHORT TWO-STRANDED ANTI-PARALLEL BETA-SHEET IS FORMED BY RESIDUES 54 - 63, ENCLOSING A TYPE II' TURN BETWEEN RESIDUES 57 AND 60. THERE ARE TWO INTERSTRAND HYDROGEN BONDS BETWEEN RESIDUES 57 AND 60. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1cyi.cif.gz 1cyi.cif.gz | 34.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1cyi.ent.gz pdb1cyi.ent.gz | 22.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1cyi.json.gz 1cyi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cy/1cyi https://data.pdbj.org/pub/pdb/validation_reports/cy/1cyi ftp://data.pdbj.org/pub/pdb/validation_reports/cy/1cyi ftp://data.pdbj.org/pub/pdb/validation_reports/cy/1cyi | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 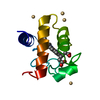
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 9810.996 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | ChemComp-CD / #3: Chemical | ChemComp-HEC / | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.39 Å3/Da / Density % sol: 48.6 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal | *PLUS Density % sol: 49.1 % | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS pH: 7.5 / Method: vapor diffusion | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.54 |
|---|---|
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: Sep 1, 1991 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 1.9→41.3 Å / Num. obs: 6412 / % possible obs: 84.8 % / Observed criterion σ(I): 0 / Redundancy: 3.03 % / Rmerge(I) obs: 0.064 |
| Reflection | *PLUS Num. measured all: 19437 / Rmerge(I) obs: 0.064 |
| Reflection shell | *PLUS Mean I/σ(I) obs: 2.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.9→10 Å / σ(F): 0 Details: NO RESTRAINTS WERE PLACED ON CADMIUM IONS DURING REFINEMENT. CONSEQUENTLY THERE ARE CLOSE CONTACTS BETWEEN SOLVENT MOLECULES AND SIDE-CHAIN ATOMS INVOLVED IN CADMIUM LIGATION.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 23.9 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.9→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller












 PDBj
PDBj












