+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bxm | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ENGINEERED BETA-CRYPTOGEIN COMPLEXED WITH ERGOSTEROL | |||||||||
 Components Components | BETA-CRYPTOGEIN | |||||||||
 Keywords Keywords | FUNGAL TOXIC ELICITOR / FUNGAL TOXIC ELICITOR MUTANT / ELICITIN / PHYTOPHTHORA / STEROL / PLANT PATHOGEN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated perturbation of host programmed cell death / symbiont-mediated killing of host cell / extracellular region Similarity search - Function | |||||||||
| Biological species |  Phytophthora cryptogea (eukaryote) Phytophthora cryptogea (eukaryote) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.15 Å MOLECULAR REPLACEMENT / Resolution: 2.15 Å | |||||||||
 Authors Authors | Boissy, G. / Brunie, S. | |||||||||
 Citation Citation |  Journal: Protein Sci. / Year: 1999 Journal: Protein Sci. / Year: 1999Title: The 2.1 A structure of an elicitin-ergosterol complex: a recent addition to the Sterol Carrier Protein family. Authors: Boissy, G. / O'Donohue, M. / Gaudemer, O. / Perez, V. / Pernollet, J.C. / Brunie, S. #1:  Journal: Structure / Year: 1996 Journal: Structure / Year: 1996Title: Crystal Structure of a Fungal Elicitor Secreted by Phytophthora Cryptogea, a Member of a Novel Class of Plant Necrotic Proteins Authors: Boissy, G. / De La Fortelle, E. / Kahn, R. / Huet, J.C. / Bricogne, G. / Pernollet, J.C. / Brunie, S. #2:  Journal: Protein Expr.Purif. / Year: 1996 Journal: Protein Expr.Purif. / Year: 1996Title: Overexpression in Pichia Pastoris and Crystallization of an Elicitor Protein Secreted by the Phytopathogenic Fungus, Phytophthora Cryptogea Authors: O'Donohue, M.J. / Boissy, G. / Huet, J.C. / Nespoulous, C. / Brunie, S. / Pernollet, J.C. #3:  Journal: J.Mol.Biol. / Year: 1993 Journal: J.Mol.Biol. / Year: 1993Title: Crystallization and Preliminary X-Ray Diffraction Studies of Beta-Cryptogein, a Toxic Elicitin Secreted by the Phytopathogenic Fungus Phytophthora Cryptogea Authors: Guilloteau, J.P. / Nespoulous, C. / Huet, J.C. / Beauvais, F. / Pernollet, J.C. / Brunie, S. | |||||||||
| History |
|
- Structure visualization
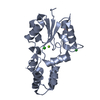
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bxm.cif.gz 1bxm.cif.gz | 33.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bxm.ent.gz pdb1bxm.ent.gz | 21.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bxm.json.gz 1bxm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bx/1bxm https://data.pdbj.org/pub/pdb/validation_reports/bx/1bxm ftp://data.pdbj.org/pub/pdb/validation_reports/bx/1bxm ftp://data.pdbj.org/pub/pdb/validation_reports/bx/1bxm | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 10492.951 Da / Num. of mol.: 1 / Mutation: T1G, A2T, K13H Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Phytophthora cryptogea (eukaryote) Phytophthora cryptogea (eukaryote)Description: N-TERMINAL ARGININE INCORPORATION NON-BIOLOGICAL SEQUENCE Cell line: PICHIA PASTORIS / Gene: CRY (MUTATED LYS 13 HIS) / Plasmid: PHILS1 / Gene (production host): CRY (MUTATED LYS 13 HIS) / Production host:  Pichia pastoris (fungus) / References: UniProt: P15570 Pichia pastoris (fungus) / References: UniProt: P15570 |
|---|---|
| #2: Chemical | ChemComp-ERG / |
| #3: Water | ChemComp-HOH / |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.3 Å3/Da / Density % sol: 47 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 4.5 / Details: pH 4.5 | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 277 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Feb 1, 1996 |
| Radiation | Monochromator: GRAPHITE(002) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→10 Å / Num. obs: 5370 / % possible obs: 96.1 % / Observed criterion σ(I): 2 / Redundancy: 6.1 % / Biso Wilson estimate: 20.9 Å2 / Rsym value: 0.046 |
| Reflection | *PLUS Num. measured all: 33143 / Rmerge(I) obs: 0.046 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: THE REFINED 2.2 ANGSTROMS RESOLUTION STRUCTURE OF THE WILD-TYPE BETA-CRYPTOGEIN (1BEO). THE ERGOSTEROL MOLECULE WAS CONSTRUCTED USING, AS A TEMPLATE, THE COORDINATES OF THE ...Starting model: THE REFINED 2.2 ANGSTROMS RESOLUTION STRUCTURE OF THE WILD-TYPE BETA-CRYPTOGEIN (1BEO). THE ERGOSTEROL MOLECULE WAS CONSTRUCTED USING, AS A TEMPLATE, THE COORDINATES OF THE CHOLESTERYL LINOLEATE MOLECULE COMPLEXED TO THE CHOLESTEROL ESTERASE (1CLE). Resolution: 2.15→8 Å / Rfactor Rfree error: 0.015 / Data cutoff high absF: 100000 / Data cutoff low absF: 0.001 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 3 / Details: RESOLUTION-DEPENDENT WEIGHTING SCHEME USED.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 18.6 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.15→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.15→2.28 Å / Rfactor Rfree error: 0.052 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller














 PDBj
PDBj


