[English] 日本語
 Yorodumi
Yorodumi- PDB-1cle: STRUCTURE OF UNCOMPLEXED AND LINOLEATE-BOUND CANDIDA CYLINDRACEA ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1cle | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF UNCOMPLEXED AND LINOLEATE-BOUND CANDIDA CYLINDRACEA CHOLESTEROL ESTERASE | |||||||||
 Components Components | CHOLESTEROL ESTERASE | |||||||||
 Keywords Keywords | LIPASE / ESTERASE / SUBSTRATE/PRODUCT-BOUND | |||||||||
| Function / homology |  Function and homology information Function and homology informationtriacylglycerol lipase / triacylglycerol lipase activity / cholesterol metabolic process / lipid catabolic process Similarity search - Function | |||||||||
| Biological species |  Candida cylindracea (fungus) Candida cylindracea (fungus) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | |||||||||
 Authors Authors | Ghosh, D. | |||||||||
 Citation Citation |  Journal: Structure / Year: 1995 Journal: Structure / Year: 1995Title: Structure of uncomplexed and linoleate-bound Candida cylindracea cholesterol esterase. Authors: Ghosh, D. / Wawrzak, Z. / Pletnev, V.Z. / Li, N. / Kaiser, R. / Pangborn, W. / Jornvall, H. / Erman, M. / Duax, W.L. #1:  Journal: FEBS Lett. / Year: 1994 Journal: FEBS Lett. / Year: 1994Title: Monomeric and Dimeric Forms of Cholesterol Esterase from Candida Cylindracea: Primary Structure, Identity in Peptide Patterns, and Additional Microheterogeneity Authors: Kaiser, R. / Erman, M. / Duax, W.L. / Ghosh, D. / Jornvall, H. #2:  Journal: J.Steroid Biochem.Mol.Biol. / Year: 1991 Journal: J.Steroid Biochem.Mol.Biol. / Year: 1991Title: Crystallization and Preliminary Diffraction Analysis of Cholesterol Esterase from Candida Cylindracea Authors: Ghosh, D. / Erman, M. / Duax, W.L. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1cle.cif.gz 1cle.cif.gz | 223.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1cle.ent.gz pdb1cle.ent.gz | 176.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1cle.json.gz 1cle.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cl/1cle https://data.pdbj.org/pub/pdb/validation_reports/cl/1cle ftp://data.pdbj.org/pub/pdb/validation_reports/cl/1cle ftp://data.pdbj.org/pub/pdb/validation_reports/cl/1cle | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: CIS PROLINE - PRO A 390 / 2: CIS PROLINE - PRO B 390 |
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 57329.469 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Candida cylindracea (fungus) / References: UniProt: P32947 Candida cylindracea (fungus) / References: UniProt: P32947 |
|---|
-Sugars , 2 types, 4 molecules 
| #2: Polysaccharide | Source method: isolated from a genetically manipulated source #3: Sugar | |
|---|
-Non-polymers , 3 types, 482 molecules 
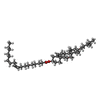



| #4: Chemical | | #5: Chemical | #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.6 Å3/Da / Density % sol: 52.75 % |
|---|---|
| Crystal grow | *PLUS Method: vapor diffusion, hanging dropDetails: please refer to McPherson, A., Preparation and Analysis of Protein Crystals. Wiley, New York (1982) |
-Data collection
| Diffraction source | Wavelength: 1.54 |
|---|---|
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: 1993 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.54 Å / Relative weight: 1 |
| Reflection | Num. obs: 119382 / % possible obs: 88 % / Observed criterion σ(I): 1 / Redundancy: 1.9 % / Rmerge(I) obs: 0.069 |
| Reflection | *PLUS Highest resolution: 2 Å / Num. obs: 62706 / Num. measured all: 119382 / Rmerge(I) obs: 0.069 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2→8 Å / σ(F): 1 /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20.5 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Classification: refinement X-PLOR / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



