+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1bgz | ||||||
|---|---|---|---|---|---|---|---|
| Title | S8 RRNA BINDING SITE FROM E. COLI, NMR, 6 STRUCTURES | ||||||
 Components Components | RNA | ||||||
 Keywords Keywords | RNA / RIBONUCLEIC ACID / 16S RRNA / INTERNAL LOOP / BASE TRIPLE | ||||||
| Function / homology | RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR / simulated annealing | ||||||
 Authors Authors | Kalurachchi, K. / Nikonowicz, E.P. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: NMR structure determination of the binding site for ribosomal protein S8 from Escherichia coli 16 S rRNA. Authors: Kalurachchi, K. / Nikonowicz, E.P. #1:  Journal: Biochim.Biophys.Acta / Year: 1979 Journal: Biochim.Biophys.Acta / Year: 1979Title: Binding Sites for Ribosomal Proteins S8 and S15 in the 16 S RNA of Escherichia Coli Authors: Zimmermann, R.A. / Singh-Bergmann, K. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1bgz.cif.gz 1bgz.cif.gz | 91.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1bgz.ent.gz pdb1bgz.ent.gz | 73.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1bgz.json.gz 1bgz.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bg/1bgz https://data.pdbj.org/pub/pdb/validation_reports/bg/1bgz ftp://data.pdbj.org/pub/pdb/validation_reports/bg/1bgz ftp://data.pdbj.org/pub/pdb/validation_reports/bg/1bgz | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 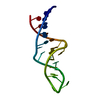
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 7356.392 Da / Num. of mol.: 1 / Fragment: 16S RRNA BINDING SITE FOR S8 / Source method: obtained synthetically / Details: RNA HAIRPIN |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: 2D AND 3D DOUBLE AND TRIPLE RESONANCE EXPERIMENTS WERE USED TO OBTAIN DISTANCE AND TORSION ANGLE CONSTRAINTS. |
- Sample preparation
Sample preparation
| Details | Contents: H2O/D2O |
|---|---|
| Sample conditions | Ionic strength: 10mM NACL, 10mM POTASSIUM PHOSPHATE / pH: 6.8 / Pressure: AMBIENT / Temperature: 298 K |
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker AMX500 / Manufacturer: Bruker / Model: AMX500 / Field strength: 500 MHz |
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: simulated annealing / Software ordinal: 1 Details: REFINEMENT DETAILS CAN BE FOUND IN THE JRNL CITATION ABOVE. | ||||||||||||
| NMR ensemble | Conformer selection criteria: FEWEST RESTRAINT VIOLATIONS / Conformers calculated total number: 50 / Conformers submitted total number: 6 |
 Movie
Movie Controller
Controller












 PDBj
PDBj





























 HSQC
HSQC