+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1awq | ||||||
|---|---|---|---|---|---|---|---|
| Title | CYPA COMPLEXED WITH HAGPIA (PSEUDO-SYMMETRIC MONOMER) | ||||||
 Components Components |
| ||||||
 Keywords Keywords | COMPLEX (ISOMERASE/PEPTIDE) / COMPLEX (ISOMERASE-PEPTIDE) / CYCLOPHILIN A / HIV-1 CAPSID / PSEUDO-SYMMETRY / COMPLEX (ISOMERASE-PEPTIDE) complex | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of protein K48-linked ubiquitination / regulation of apoptotic signaling pathway / cell adhesion molecule production / lipid droplet organization / negative regulation of viral life cycle / heparan sulfate binding / regulation of viral genome replication / virion binding / leukocyte chemotaxis / negative regulation of stress-activated MAPK cascade ...negative regulation of protein K48-linked ubiquitination / regulation of apoptotic signaling pathway / cell adhesion molecule production / lipid droplet organization / negative regulation of viral life cycle / heparan sulfate binding / regulation of viral genome replication / virion binding / leukocyte chemotaxis / negative regulation of stress-activated MAPK cascade / endothelial cell activation / Basigin interactions / protein peptidyl-prolyl isomerization / cyclosporin A binding / Minus-strand DNA synthesis / Plus-strand DNA synthesis / Uncoating of the HIV Virion / Early Phase of HIV Life Cycle / Integration of provirus / negative regulation of protein phosphorylation / APOBEC3G mediated resistance to HIV-1 infection / viral release from host cell / Calcineurin activates NFAT / activation of protein kinase B activity / Binding and entry of HIV virion / positive regulation of viral genome replication / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / negative regulation of protein kinase activity / neutrophil chemotaxis / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / Assembly Of The HIV Virion / positive regulation of protein secretion / Budding and maturation of HIV virion / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / positive regulation of NF-kappaB transcription factor activity / platelet activation / platelet aggregation / SARS-CoV-1 activates/modulates innate immune responses / integrin binding / positive regulation of protein phosphorylation / neuron differentiation / unfolded protein binding / Platelet degranulation / protein folding / cellular response to oxidative stress / vesicle / secretory granule lumen / ficolin-1-rich granule lumen / positive regulation of MAPK cascade / focal adhesion / apoptotic process / Neutrophil degranulation / protein-containing complex / extracellular space / RNA binding / extracellular exosome / extracellular region / nucleus / membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.58 Å MOLECULAR REPLACEMENT / Resolution: 1.58 Å | ||||||
 Authors Authors | Vajdos, F.F. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 1997 Journal: Protein Sci. / Year: 1997Title: Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Authors: Vajdos, F.F. / Yoo, S. / Houseweart, M. / Sundquist, W.I. / Hill, C.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1awq.cif.gz 1awq.cif.gz | 45.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1awq.ent.gz pdb1awq.ent.gz | 32.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1awq.json.gz 1awq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  1awq_validation.pdf.gz 1awq_validation.pdf.gz | 430.8 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  1awq_full_validation.pdf.gz 1awq_full_validation.pdf.gz | 435.5 KB | Display | |
| Data in XML |  1awq_validation.xml.gz 1awq_validation.xml.gz | 9.3 KB | Display | |
| Data in CIF |  1awq_validation.cif.gz 1awq_validation.cif.gz | 11.8 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/aw/1awq https://data.pdbj.org/pub/pdb/validation_reports/aw/1awq ftp://data.pdbj.org/pub/pdb/validation_reports/aw/1awq ftp://data.pdbj.org/pub/pdb/validation_reports/aw/1awq | HTTPS FTP |
-Related structure data
| Related structure data |  1awrC  1awsC  1awtC  1awuC  1awvC  2cyhS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 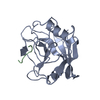
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 17905.307 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Cellular location: CYTOPLASM / Gene: CYCLOPHILIN / Gene (production host): CYCLOPHILIN / Production host: Homo sapiens (human) / Cellular location: CYTOPLASM / Gene: CYCLOPHILIN / Gene (production host): CYCLOPHILIN / Production host:  |
|---|---|
| #2: Protein/peptide | Mass: 565.642 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.25 Å3/Da / Density % sol: 45.42 % | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 8.4 / Details: pH 8.4 | ||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 21 ℃ / pH: 8.2 / Method: unknown | ||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X12C / Wavelength: 1.1 / Beamline: X12C / Wavelength: 1.1 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Oct 28, 1995 |
| Radiation | Monochromator: SI(111) / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.58→15 Å / Num. obs: 24895 / % possible obs: 88.1 % / Observed criterion σ(I): 0 / Biso Wilson estimate: 20 Å2 / Rsym value: 0.086 / Net I/σ(I): 9.4 |
| Reflection shell | Resolution: 1.58→1.61 Å / Rsym value: 0.156 / % possible all: 51.9 |
| Reflection | *PLUS Num. measured all: 214067 / Rmerge(I) obs: 0.086 |
| Reflection shell | *PLUS % possible obs: 51.9 % / Rmerge(I) obs: 0.156 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 2CYH Resolution: 1.58→15 Å / Rfactor Rfree error: 0.014 / Data cutoff high absF: 10000000 / Data cutoff low absF: 0.001 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 Details: BULK SOLVENT MODEL USED REGARDING THE HIGH R-VALUE: THIS IS THE STRUCTURE OF THE COMPLEX REFINED IN A PSEUDO-SPACE GROUP. THE DETAILS ARE ADDRESSED EXTENSIVELY IN THE PAPER. TO SUMMARIZE ...Details: BULK SOLVENT MODEL USED REGARDING THE HIGH R-VALUE: THIS IS THE STRUCTURE OF THE COMPLEX REFINED IN A PSEUDO-SPACE GROUP. THE DETAILS ARE ADDRESSED EXTENSIVELY IN THE PAPER. TO SUMMARIZE HERE, THE HIGH R-VALUE FOR THIS STRUCTURE STEMS FROM THE BREAKDOWN IN PSEUDO-SYMMETRY AT HIGH RESOLUTION.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 27.2 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.58→15 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.58→1.68 Å / Rfactor Rfree error: 0.05 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.843 / Classification: refinement X-PLOR / Version: 3.843 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller












 PDBj
PDBj












