[English] 日本語
 Yorodumi
Yorodumi- PDB-149d: SOLUTION STRUCTURE OF A PYRIMIDINE(DOT)PURINE(DOT) PYRIMIDINE DNA... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 149d | ||||||
|---|---|---|---|---|---|---|---|
| Title | SOLUTION STRUCTURE OF A PYRIMIDINE(DOT)PURINE(DOT) PYRIMIDINE DNA TRIPLEX CONTAINING T(DOT)AT, C+(DOT)GC AND G(DOT)TA TRIPLES | ||||||
 Components Components |
| ||||||
 Keywords Keywords | DNA / TRIPLEX | ||||||
| Function / homology | DNA Function and homology information Function and homology information | ||||||
| Method | SOLUTION NMR | ||||||
 Authors Authors | Radhakrishnan, I. / Patel, D.J. | ||||||
 Citation Citation |  Journal: Structure / Year: 1994 Journal: Structure / Year: 1994Title: Solution structure of a pyrimidine.purine.pyrimidine DNA triplex containing T.AT, C+.GC and G.TA triples. Authors: Radhakrishnan, I. / Patel, D.J. #1:  Journal: Biochemistry / Year: 1992 Journal: Biochemistry / Year: 1992Title: Three-Dimensional Homonuclear Noesy-Tocsy of an Intramolecular Pyrimidine(Dot)Purine(Dot)Pyrimidine DNA Triplex Containing a Central G(Dot)Ta Triple: Nonexchangeable Proton Assignments and Structural Implications Authors: Radhakrishnan, I. / Patel, D.J. / Gao, X. #2:  Journal: J.Am.Chem.Soc. / Year: 1992 Journal: J.Am.Chem.Soc. / Year: 1992Title: Solution Conformation of a G(Dot)Ta Triple in an Intramolecular Pyrimidine(Dot)Purine(Dot)Pyrimidine DNA Triplex Authors: Radhakrishnan, I. / Patel, D.J. / Veal, J.M. / Gao, X. #3:  Journal: Biochemistry / Year: 1991 Journal: Biochemistry / Year: 1991Title: NMR Structural Studies of Intramolecular (Y+)N(Dot)(R+)N(Y-)N DNA Triplexes in Solution: Imino and Amino Proton and Nitrogen Markers of G(Dot)Ta Base Triple Formation Authors: Radhakrishnan, I. / Gao, X. / De Los Santos, C. / Live, D. / Patel, D.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  149d.cif.gz 149d.cif.gz | 218.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb149d.ent.gz pdb149d.ent.gz | 153.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  149d.json.gz 149d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/49/149d https://data.pdbj.org/pub/pdb/validation_reports/49/149d ftp://data.pdbj.org/pub/pdb/validation_reports/49/149d ftp://data.pdbj.org/pub/pdb/validation_reports/49/149d | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 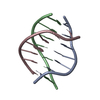
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: THREE RESIDUES IN THE SEQUENCE STUDIED ARE CYTOSINES THAT ARE PROTONATED AT THE N3 POSITION. THESE RESIDUES DIFFER FROM "NORMAL" CYTOSINES IN THAT THEY HAVE A PROTON ATTACHED TO THE N3 NITROGEN. ...1: THREE RESIDUES IN THE SEQUENCE STUDIED ARE CYTOSINES THAT ARE PROTONATED AT THE N3 POSITION. THESE RESIDUES DIFFER FROM "NORMAL" CYTOSINES IN THAT THEY HAVE A PROTON ATTACHED TO THE N3 NITROGEN. IT SHOULD BE NOTED THAT THIS IS NOT A PERMANENT MODIFICATION BUT RATHER A CONSEQUENCE OF THE CONDITIONS UNDER WHICH THE SEQUENCE WAS STUDIED. THESE N3-PROTONATED CYTOSINE RESIDUES HAVE BEEN ASSIGNED THE RESIDUE NAME C IN THE ENTRY AND ATOM H3 HAS BEEN FOOTNOTED. | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain | Mass: 2048.373 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: CHEMICALLY SYNTHESIZED |
|---|---|
| #2: DNA chain | Mass: 2186.473 Da / Num. of mol.: 1 / Source method: obtained synthetically |
| #3: DNA chain | Mass: 2064.372 Da / Num. of mol.: 1 / Source method: obtained synthetically |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
| Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name:  X-PLOR / Developer: BRUNGER / Classification: refinement X-PLOR / Developer: BRUNGER / Classification: refinement | ||||||||
| Refinement | Software ordinal: 1 Details: R VALUE 0.022 RMSD BOND DISTANCES 0.005 ANGSTROMS RMSD BOND ANGLES 2.26 DEGREES | ||||||||
| NMR ensemble | Conformers submitted total number: 14 |
 Movie
Movie Controller
Controller









 PDBj
PDBj

